AthCNV: A map of DNA copy number variations in the Arabidopsis thaliana genome (Plant Cell)
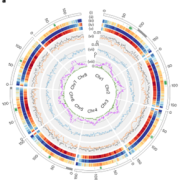
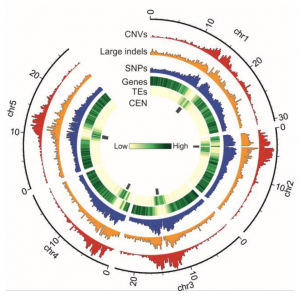 Intraspecific phenotypic variability can be attributed to differences at the genome level such as copy number variations (CNVs). CNVs are large DNA fragments which differ in number between individuals and likely have a crucial evolutionary role. Due to the 1001 Genomes Project, over 1,000 Arabidopsis accessions have been sequenced, with to-date analyses largely focusing on single nucleotide polymorphisms (SNPs) and other small variations. However, there is limited understanding regarding the extent of CNV contribution to genotypic (and phenotypic) diversity. Here, Zmienko et al. generated AthCNV, a map of CNVs in Arabidopsis, which has substantially added to the list of known structural variants. The authors found that CNVs constitute one-third of the genome and are unevenly distributed throughout, with transposable elements overrepresented in these regions. 18.3% of genes fully overlap with CNVs and this group is enriched with genes functioning in defense and stress. To investigate the Arabidopsis population structure, principal component analyses were conducted using CNV markers. Interestingly, the CNV-approach better determined the global distribution and migration patterns of accessions than SNPs. Furthermore, gene copy number losses were more frequent than gains, and 3.9-26.9% of CNV-affected genes were implicated by gene copy number changes in individual accessions. Finally, to link CNVs to phenotypic variation, CNV markers were utilized to perform a genome-wide association study and a strong association was found between Pseudomonas resistance and two CNV-affected R genes. AthCNV has revealed a multitude of previously undetected genomic variants, and this map will facilitate future research to illuminate the impact of CNVs on phenotypic variability in Arabidopsis. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00640
Intraspecific phenotypic variability can be attributed to differences at the genome level such as copy number variations (CNVs). CNVs are large DNA fragments which differ in number between individuals and likely have a crucial evolutionary role. Due to the 1001 Genomes Project, over 1,000 Arabidopsis accessions have been sequenced, with to-date analyses largely focusing on single nucleotide polymorphisms (SNPs) and other small variations. However, there is limited understanding regarding the extent of CNV contribution to genotypic (and phenotypic) diversity. Here, Zmienko et al. generated AthCNV, a map of CNVs in Arabidopsis, which has substantially added to the list of known structural variants. The authors found that CNVs constitute one-third of the genome and are unevenly distributed throughout, with transposable elements overrepresented in these regions. 18.3% of genes fully overlap with CNVs and this group is enriched with genes functioning in defense and stress. To investigate the Arabidopsis population structure, principal component analyses were conducted using CNV markers. Interestingly, the CNV-approach better determined the global distribution and migration patterns of accessions than SNPs. Furthermore, gene copy number losses were more frequent than gains, and 3.9-26.9% of CNV-affected genes were implicated by gene copy number changes in individual accessions. Finally, to link CNVs to phenotypic variation, CNV markers were utilized to perform a genome-wide association study and a strong association was found between Pseudomonas resistance and two CNV-affected R genes. AthCNV has revealed a multitude of previously undetected genomic variants, and this map will facilitate future research to illuminate the impact of CNVs on phenotypic variability in Arabidopsis. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00640