An optimized high-throughput assay for the identification of enhancers in plant genomes
Jores et al. adapt STARR-seq for identifying enhancers in tobacco leaves.
Plant Cell https://doi.org/10.1105/tpc.20.00155
By Tobias Joresa, Jackson Tonniesa,b, Michael W Dorritya, Josh T Cuperusa, Stanley Fieldsa,c and Christine Queitscha
a Department of Genome Sciences, University of Washington, Seattle, WA 98195.
b Graduate Program in Biology, University of Washington, Seattle, WA 98195.
c Department of Medicine, University of Washington, Seattle, WA 98195.
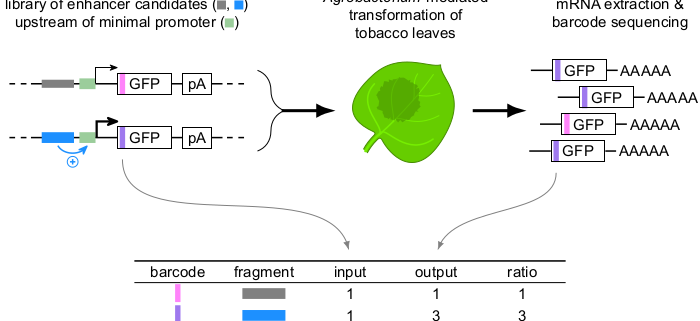
Background: Precise regulation of gene expression is vital for development and adaptation to the environment. Relatively short DNA sequences known as enhancers play a key role in gene regulation by increasing the amount of transcription from associated genes, often in a condition- or tissue-specific manner. Enhancers can be located up- or downstream of their target genes and up to several kilobases away. This flexibility in enhancer location makes it hard to find enhancers. In 2013, a high-throughput assay termed STARR-seq was developed to identify enhancers by inserting candidate sequences into a reporter gene such that they become present in the mRNA for detection. In plants, this assay has suffered from a relatively low signal-to-noise ratio.
Question: We set out to adapt STARR-seq for its use in tobacco leaves and to optimize the assay to increase its dynamic range.
Findings: We established a variant of the STARR-seq assay that works in tobacco leaves and enables precise and reproducible detection of enhancer activity. We found that the location of enhancers within the reporter construct has a strong influence on their activity. Enhancers were most active immediately upstream of the promoter. By contrast, enhancers showed no activity when they were part of the transcribed region of the reporter gene. This version of the STARR-seq assay can detect condition-specific enhancers (response to light) and can pinpoint underlying functional elements.
Next steps: In future work, the assay that we established here can be used to identify enhancers globally within plant genomes. The results will help to improve our understanding of these key gene-regulatory elements.
Tobias Jores, Jackson Tonnies, Michael W Dorrity, Josh T Cuperus, Stanley Fields and Christine Queitsch. (2020). Identification of Plant Enhancers and Their Constituent Elements by STARR-seq in Tobacco Leaves. Plant Cell; DOI: https://doi.org/10.1105/tpc.20.00155