An interactome of plant autophagy proteins and pathogen effectors (Cell Host Microbe) ($)
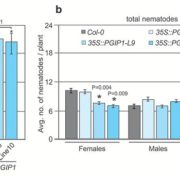
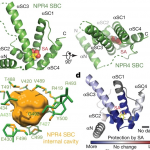
 Autophagy is a regulated process whereby select cytoplasmic components are degraded. It is involved in a variety of biological processes, including defense against pathogens. As such, plant pathogens have evolved mechanisms to target host autophagy machinery to gain virulence. However, we still lack a comprehensive understanding of pathogen molecules that alter plant autophagy pathways. Lal et al. conducted a yeast two-hybrid screen between 25 Arabidopsis autophagy (ATG) proteins and 184 effectors derived from diverse pathogens and found 88 interacting pairs. Among ATG proteins, ATG8 interacted with the highest number of effectors. ATG8 is a central player in autophagy processes such as the promotion of autophagosome biogenesis and the recruitment of specific cargo for degradation. The authors characterized the functions of three effectors of the bacterial pathogen Pseudomonas syringae pv tomato, HrpZ1, HopF1, and AvrPtoB. HrpZ1 and HopF1 both interact with ATG8 but regulate autophagy in opposite directions. Heterologous expression of HrpZ1 or HopF1 in plants conferred susceptibility to a bacterial pathogen. AvrPtoB does not interact with ATG8 but with ATG1, another frequently targeted ATG protein. The authors showed AvrPtoB inhibits autophosphorylation of ATG1, affecting its kinase activity. This study provides a resource that helps researchers to study molecular mechanisms by which pathogens target autophagy processes of plants. (Summary by Tatsuya Nobori @nobolly) Cell Host Microbe 10.1016/j.chom.2020.07.010
Autophagy is a regulated process whereby select cytoplasmic components are degraded. It is involved in a variety of biological processes, including defense against pathogens. As such, plant pathogens have evolved mechanisms to target host autophagy machinery to gain virulence. However, we still lack a comprehensive understanding of pathogen molecules that alter plant autophagy pathways. Lal et al. conducted a yeast two-hybrid screen between 25 Arabidopsis autophagy (ATG) proteins and 184 effectors derived from diverse pathogens and found 88 interacting pairs. Among ATG proteins, ATG8 interacted with the highest number of effectors. ATG8 is a central player in autophagy processes such as the promotion of autophagosome biogenesis and the recruitment of specific cargo for degradation. The authors characterized the functions of three effectors of the bacterial pathogen Pseudomonas syringae pv tomato, HrpZ1, HopF1, and AvrPtoB. HrpZ1 and HopF1 both interact with ATG8 but regulate autophagy in opposite directions. Heterologous expression of HrpZ1 or HopF1 in plants conferred susceptibility to a bacterial pathogen. AvrPtoB does not interact with ATG8 but with ATG1, another frequently targeted ATG protein. The authors showed AvrPtoB inhibits autophosphorylation of ATG1, affecting its kinase activity. This study provides a resource that helps researchers to study molecular mechanisms by which pathogens target autophagy processes of plants. (Summary by Tatsuya Nobori @nobolly) Cell Host Microbe 10.1016/j.chom.2020.07.010