ALOG transcription factors influence the morphological diversity of plant lateral organs (bioRxiv)
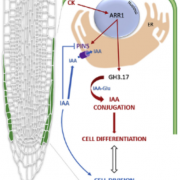
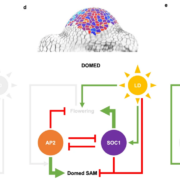
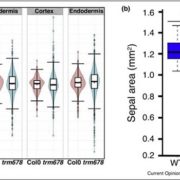
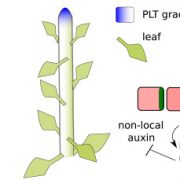
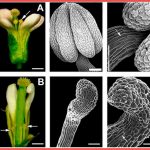
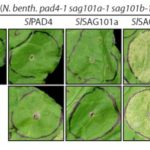
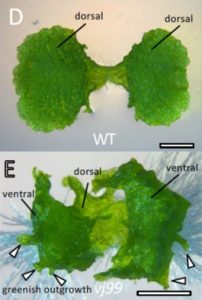 Land plants evolved from freshwater streptophytic algae over 450 million years ago and have since separated into morphologically diverse lineages. A key feature in the transition from aquatic to terrestrial life was the development of 3D body plans with lateral organs. In a new preprint, Naramoto and colleagues describe the identification of an evolutionarily conserved ALOG (Arabidopsis LSH1 and Oryza G1) family protein that regulates meristem identity and lateral organ development in an early-divergent model system. Forward genetic screening in the thalloid liverwort Marchantia polymorpha identified mutants of the ALOG gene MpTAWAWA (Mptaw) that develop numerous outgrowths from the margins of thalli relative to wild-type plants. These outgrowths resemble ventral scales that normally appear on the underside of liverworts and help anchor the plant body to the substrate. Intriguingly, Mptaw mutant thalli lack ventral scales. Furthermore, in Mptaw apical meristems show a dispersed notch-like area relative to wild-type. An apical meristem-associated marker gene expression (YUC2A::GUS) revealed that this dispersed notch-like area is indeed meristem-like in identity and is likely caused by a lack of cell division that would normally separate bifurcating meristems. In Oryza sativa (rice), the ALOG protein G1 is involved in the development of sterile lemmas (a small leaf-like lateral organ present on the rice spikelet) such that g1 mutants display abnormally enlarged lemmas. Excitingly, the ectopic expression of MpTAW in g1 mutants rescued this lateral organ defect, demonstrating functional conservation in ALOG function between these two phylogenetically-distant plant lineages. Collectively, this work underscores the power of such macroevolutionary evo-devo analyses and identifies ALOG transcription factors as critical regulators of lateral organ development in plants. (Summary by Phil Carella) bioRxiv
Land plants evolved from freshwater streptophytic algae over 450 million years ago and have since separated into morphologically diverse lineages. A key feature in the transition from aquatic to terrestrial life was the development of 3D body plans with lateral organs. In a new preprint, Naramoto and colleagues describe the identification of an evolutionarily conserved ALOG (Arabidopsis LSH1 and Oryza G1) family protein that regulates meristem identity and lateral organ development in an early-divergent model system. Forward genetic screening in the thalloid liverwort Marchantia polymorpha identified mutants of the ALOG gene MpTAWAWA (Mptaw) that develop numerous outgrowths from the margins of thalli relative to wild-type plants. These outgrowths resemble ventral scales that normally appear on the underside of liverworts and help anchor the plant body to the substrate. Intriguingly, Mptaw mutant thalli lack ventral scales. Furthermore, in Mptaw apical meristems show a dispersed notch-like area relative to wild-type. An apical meristem-associated marker gene expression (YUC2A::GUS) revealed that this dispersed notch-like area is indeed meristem-like in identity and is likely caused by a lack of cell division that would normally separate bifurcating meristems. In Oryza sativa (rice), the ALOG protein G1 is involved in the development of sterile lemmas (a small leaf-like lateral organ present on the rice spikelet) such that g1 mutants display abnormally enlarged lemmas. Excitingly, the ectopic expression of MpTAW in g1 mutants rescued this lateral organ defect, demonstrating functional conservation in ALOG function between these two phylogenetically-distant plant lineages. Collectively, this work underscores the power of such macroevolutionary evo-devo analyses and identifies ALOG transcription factors as critical regulators of lateral organ development in plants. (Summary by Phil Carella) bioRxiv