ADP-ribosylation and plant immunity
Minsoo Yoon, The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
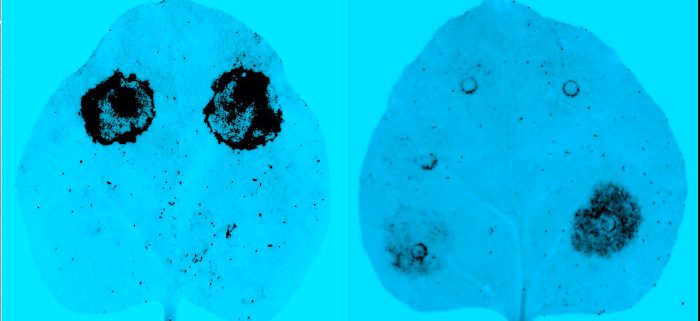
Background: Gram-negative bacterial phytopathogens inject specialized proteins (effectors) into their hosts to neutralize defense and manipulate host metabolism for the pathogen’s survival. Defense responses are initiated when host resistance (R) proteins recognize translocated bacterial effectors. Effector AvrRpm1 functions as an ADP-ribosyl transferase modifying RESISTANCE TO P. SYRINGAE PV MACULICOLA1 (RPM1)-INTERACTING PROTEIN4 (RIN4), leading to the activation of Arabidopsis protein RPM1. Another effector, AvrB, is also recognized in Arabidopsis such that RPM1 activation is linked to the phosphorylation of a conserved threonine (T166) in RIN4. A previous study proposed that ADP-ribosylation of RIN4 by AvrRpm1 leads to the RPM1-mediated defense response by inducing phosphorylation of T166.
Question: The two post-translational modifications in RIN4 (direct ADP-ribosylation by AvrRpm1 and induced phosphorylation by AvrB) are biochemically distinct, and their roles in pathogen virulence or host immunity are unclear. We identified ADP-ribosyl transferase activity of AvrRpm2 from another Pseudomonas strain and investigated whether ADP-ribosylation or RPM1 activation is affected by T166.
Findings: We conducted mutational analysis in combination with mass spectrometry to locate the target site. Only one conserved glutamate residue (E156) can be ADP-ribosylated to activate RPM1 among candidate target residues identified from MS/MS fragmentation spectra. Soybean and snap bean RIN4 homologs without glutamate at the positions corresponding to the E156 of Arabidopsis RIN4 are not ADP-ribosylated by bacterial AvrRpm2. In contrast with effector AvrB, AvrRpm2 does not require the phosphorylation of T166 to activate RPM1. This study suggests that a common resistance protein can recognize separate biochemical reactions by different pathogen effectors through distinct modifications of RIN4.
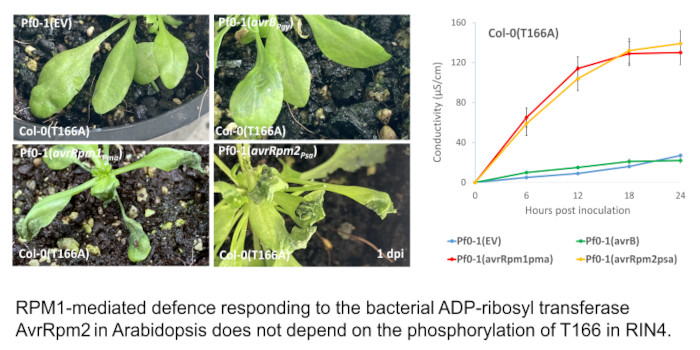
Next steps: We confirmed that AvrRpm2 contributes to pathogen virulence. It would be interesting to see how the virulence function of the pathogen effector is related to the ADP-ribosylation of the host protein RIN4.
Reference:
Minsoo Yoon, Martin J. Middleditch, Erik H. A. Rikkerink (2022). A conserved glutamate residue in RPM1-INTERACTING PROTEIN4 is ADP-ribosylated by the Pseudomonas effector AvrRpm2 to activate RPM1-mediated plant resistance. Plant Cell. https://doi.org/10.1093/plcell/koac286