A small nucleolar RNA cluster that directs ribosomal RNA maturation
Cao et al. found that SnoR28.1 (a conserved box C/D snoRNA cluster) regulates plant growth and development by directing rRNA maturation. The Plant Cell (2022). https://doi.org/10.1093/plcell/koac265
By Yuxin Cao and Weiqiang Qian
Background: Small nucleolar RNAs (snoRNAs) are a class of abundantly expressed noncoding RNAs that guide modifications of ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and small nuclear RNAs (snRNAs) within the nucleolus. In plants, many snoRNAs have been identified and annotated. However, the biological functions of most snoRNAs in higher plants remain unclear. Based on sequences of conserved elements, snoRNAs are classified into box C/D or box H/ACA snoRNAs, which catalyze 2′-O-ribose methylation and pseudouridinylation, respectively. SnoRNAs associate with different sets of core proteins to form small nucleolar ribonucleoprotein particles. For rRNA modification, they base-pair with complementary sequences on rRNA precursors to achieve site-specificity. In addition to rRNA modification, a few snoRNAs are required for proper and efficient pre-rRNA processing.
Question: What is the function of Arabidopsis thaliana snoRNAs whose functions remain to be characterized? Can we use a reverse genetic approach to identify snoRNAs with important functions in plant development?
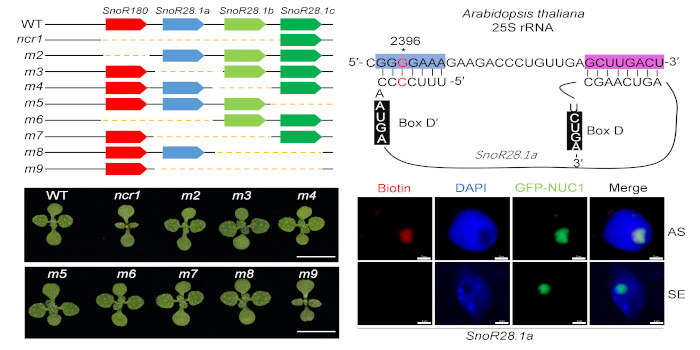
Findings: By screening a library of CRISPR/Cas9-generated noncoding RNA deletion mutants, we identified SnoR28.1, a locus containing a cluster of genes producing conserved box C/D snoRNAs, as an important regulator of plant growth and development. Deletion of the SnoR28.1 cluster results in pleiotropic root and shoot growth defects. SnoR28.1s guide 2′-O-ribose methylation of 25S rRNA G2396 and facilitate pre-rRNA processing, which is essential for proper ribosome function. SnoR28 contains a 7-bp antisense box (shortest) and an 8-bp ‘extra box’ that is complementary to the rRNA methylation site nearby. The antisense box is required for 25S rRNA methylation and pre-rRNA processing. The ‘extra box’ is also required for pre-rRNA processing, but is partially responsible for methylation. Our results provide insight into the biological role of a snoRNA cluster in plants.
 Next steps: We are using ribosome profiling to identify proteins whose translation is affected by loss of SnoR28.1s. The biological importance of other snoRNAs in plants also remains an exciting question for future research.
Next steps: We are using ribosome profiling to identify proteins whose translation is affected by loss of SnoR28.1s. The biological importance of other snoRNAs in plants also remains an exciting question for future research.
Reference:
Yuxin Cao, Jiayin Wang, Songlin Wu, Xiaochang Yin, Jia Shu, Xing Dai, Yannan Liu, Linhua Sun, Danmeng Zhu, Xing Wang Deng, Keqiong Ye and Weiqiang Qian (2022). The small nucleolar RNA SnoR28 regulates plant growth and development by directing rRNA maturation. Plant Cell. https://doi.org/10.1093/plcell/koac265



