A revised model for purine nucleotide catabolism
Baccolini and Witte present a revised model for nucleotide catabolism in Arabidopsis. Plant Cell https://doi.org/10.1105/tpc.18.00899
Chiara Baccolini, Leibniz Universität Hannover
Prof. Dr. Claus-Peter Witte, Leibniz Universität Hannover
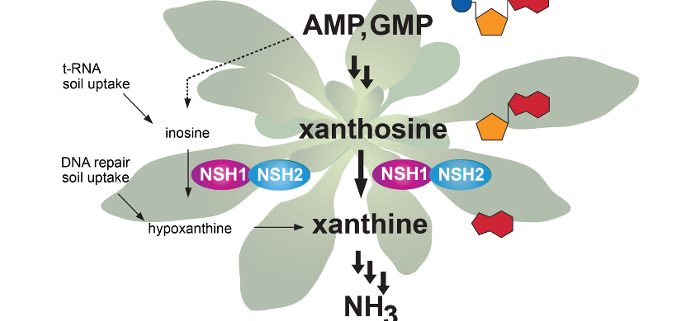
Background: Nitrogen is a key element for plant growth and is constantly recycled in plant metabolism, for example, by the breakdown of the nitrogen-containing bases in nucleic acids. The current model of AMP and GMP degradation proposes a branched network of reactions, either involving inosine and hypoxanthine or alternatively xanthosine as intermediates to reach the nucleobase xanthine, which is the established entry point for the reactions that release nitrogen from the purine ring. A central enzyme for both branches is nucleoside hydrolase 1 (NSH1), which hydrolyzes both inosine and xanthosine. NSH1 is also required in pyrimidine nucleotide degradation for the hydrolysis of uridine. Apart from NSH1, plants possess a conserved second nucleoside hydrolase, NSH2, of unclear function.
Question: Is the current model of purine nucleotide breakdown correct? Are the inosine and xanthosine routes both employed for the degradation of AMP and GMP? Is NSH2 involved in purine nucleotide degradation and what is its role?
Findings: Quantifying metabolites by mass spectrometry in a series of Arabidopsis mutants and double mutants of genes involved in purine nucleotide degradation, we show that AMP and GMP are mainly degraded via xanthosine, whereas the route via inosine is not used. However, inosine and hypoxanthine were always present in small amounts, probably derived from other sources, such as vacuolar t-RNA breakdown or DNA repair. By biochemical analysis, we show that NSH2 forms a complex with NSH1 and is activated by this interaction. The NSH1/NSH2 complex is a highly efficient xanthosine (and inosine) hydrolase, whereas NSH1 alone is a more efficient uridine hydrolase. We conclude that Arabidopsis and probably many other plants degrade AMP and GMP via xanthosine as intermediate employing an NSH1/NSH2 complex for xanthosine hydrolysis.
Next steps: Which are the enzymatic steps leading to the production of xanthosine from AMP and GMP, in particular which are the phosphatases involved? How is the entry into purine nucleotide degradation regulated, and how is the AMP degradation route functionally separated from the biosynthetic route of GMP, since both routes share common intermediates?
Chiara Baccolini, Claus-Peter Witte (2018). AMP and GMP Catabolism in Arabidopsis Converge on Xanthosine, which Is Degraded by a Nucleoside Hydrolase Heterocomplex. Plant Cell; DOI: https://doi.org/10.1105/tpc.18.00899
Key words: Nucleotide metabolism, protein-protein interaction, nucleoside hydrolase, mass spectrometry.