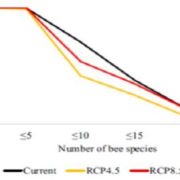
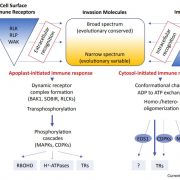
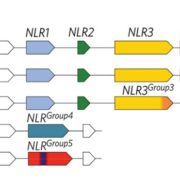
A plant secondary metabolite selectively targets bacterial pathogenicity (Cell Host Microbe)
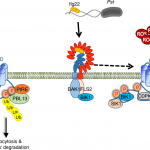
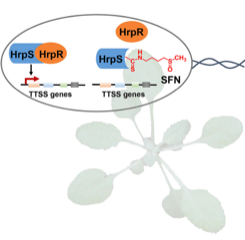 Plants have a variety of secondary metabolites that are associated with defense against pathogens, but the mode of action of such metabolites is poorly understood. Wang et al. revealed that the Arabidopsis secondary metabolite sulforaphane (SFN), which is derived from aliphatic glucosinolate, suppresses transcript accumulation of genes involved in the type III secretion system (T3SS), an essential virulence component of the bacterial pathogen Pseudomonas syringae. SFN accumulates in the leaf apoplast, site of P. syringae colonization, in a manner dependent on MYB28 and MYB29 transcription factors. In myb28myb29 mutant plants, exogenous application of SFN at the physiological concentration was sufficient to suppress T3SS expression and bacterial growth. SFN did not affect bacterial growth in vitro at the physiological concentration, and the leaf microbiota of the myb28myb29 plants grown in natural soil resembled that of wild type plants, suggesting that SFN does not function as a general antimicrobial in plants. The authors identified that SFN preferentially reacts with Cys209 of HrpS, an important regulator of the T3SS. Further, they showed that HrpSCys209 is required for HrpS to induce the T3SS and promote bacterial growth in planta. Interestingly, HrpSCys209 is highly conserved in P. syringae isolates non-adapted to Arabidopsis, but not in adapted isolates, raising a possibility that targeting of HrpS by SFN may pose a selective pressure on the evolution of plant-associated bacteria. This study provides mechanistic insights into an understudied but vital question: “how do plants affect pathogens to suppress their growth?” (Summary by Tatsuya Nobori @nobolly) Cell Host & Microbe 10.1016/j.chom.2020.03.004
Plants have a variety of secondary metabolites that are associated with defense against pathogens, but the mode of action of such metabolites is poorly understood. Wang et al. revealed that the Arabidopsis secondary metabolite sulforaphane (SFN), which is derived from aliphatic glucosinolate, suppresses transcript accumulation of genes involved in the type III secretion system (T3SS), an essential virulence component of the bacterial pathogen Pseudomonas syringae. SFN accumulates in the leaf apoplast, site of P. syringae colonization, in a manner dependent on MYB28 and MYB29 transcription factors. In myb28myb29 mutant plants, exogenous application of SFN at the physiological concentration was sufficient to suppress T3SS expression and bacterial growth. SFN did not affect bacterial growth in vitro at the physiological concentration, and the leaf microbiota of the myb28myb29 plants grown in natural soil resembled that of wild type plants, suggesting that SFN does not function as a general antimicrobial in plants. The authors identified that SFN preferentially reacts with Cys209 of HrpS, an important regulator of the T3SS. Further, they showed that HrpSCys209 is required for HrpS to induce the T3SS and promote bacterial growth in planta. Interestingly, HrpSCys209 is highly conserved in P. syringae isolates non-adapted to Arabidopsis, but not in adapted isolates, raising a possibility that targeting of HrpS by SFN may pose a selective pressure on the evolution of plant-associated bacteria. This study provides mechanistic insights into an understudied but vital question: “how do plants affect pathogens to suppress their growth?” (Summary by Tatsuya Nobori @nobolly) Cell Host & Microbe 10.1016/j.chom.2020.03.004
[altmetric doi=”10.1016/j.chom.2020.03.004″ details=”right” float=”right”]