A Genome-wide Approach to Understanding a Non-Canonical ARF
The canonical auxin-response pathway in plants begins with auxin sensing by F-box proteins, triggering degradation of AUX/IAA proteins that act as transcriptional repressors via their interaction with sequence-specific DNA-binding AUXIN RESPONSE FACTORS (ARFs; reviewed in Weijers and Wagner, 2016). Recently, the unusual ARF protein ETTIN (ETT) was proposed to represent an even shorter pathway through which plants can effect transcriptional changes in response to auxin: ETT is a transcription factor that itself shows auxin-responsive binding to protein partners, suggesting that it both senses auxin and regulates target genes (Simonini et al., 2016). Now, Simonini et al. (2017) have generated several genome-wide data sets to elegantly explore how this non-canonical ARF functions in floral organ development.
Simonini and coauthors used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to identify ETT-bound regions in the genome, which represent candidate loci to be direct targets of the ETT transcription factor. Next, the authors compared RNA-seq data sets from inflorescence tissues of the ett-3 mutant and wild type to identify floral development genes with expression regulated by ETT. The intersection of these two data sets revealed 663 candidate direct target genes whose correct expression depends on ETT.
ETT upregulated some genes, but downregulated others, showing a trend of repressing auxin signaling-related targets, but upregulating ethylene- and jasmonic acid-related targets. In addition, direct targets of ETT included genes for auxin influx and efflux carriers, as well as auxin biosynthetic genes. Indeed, Simonini et al. found that the distribution of auxin carriers, and of signals from auxin reporters, was abnormal in the ett mutant background. These data highlight the role of ETT as a hub for hormone signaling in development and suggest that ETT influences the development of floral organs via auxin biosynthesis, transport, and signaling. As ETT expression is itself regulated by auxin, these findings point to a feedback loop in the relationship between auxin and ETT.
The authors explored ETT’s role in auxin responses using data sets generated from IAA-treated plants. Notably, the overlap between the ChIP-seq data prepared with and without IAA revealed 1520 loci displaying auxin-independent ETT interactions. Another 375 loci were bound by ETT specifically without IAA, and 104 were bound specifically in the presence of IAA. Among these 479 auxin-dependent interactions, 136 genes were regulated by ETT (i.e., their expression was altered in the ett mutant compared to wild type, see Figure 1). The authors further checked whether ETT mediates auxin-responsive expression of these 136 genes by asking whether they showed auxin-responsive expression in the wild-type but not in the ett mutant. In all, the authors used comparisons of their multiple data sets to identify 77 genes that were bound by ETT in an auxin-sensitive manner, had their expression regulated by ETT and for which responsiveness of expression to IAA required ETT.
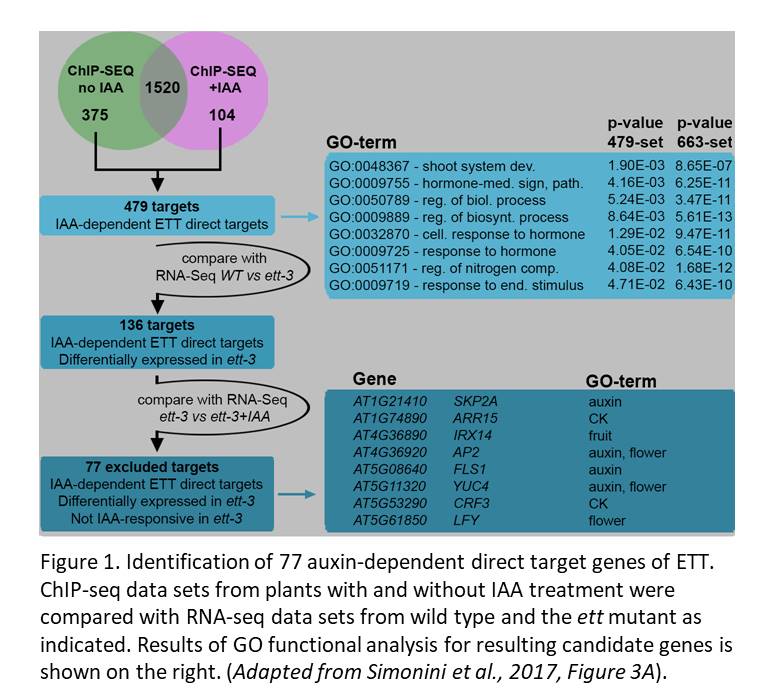 To demonstrate the usefulness of this set of auxin-responsive ETT direct target candidates, the authors explored the regulation of a few in more detail. In light of the previously proposed model that IAA prompts dissociation of ETT from a partner that mediates binding to a specific gene locus, they made use of available ChIP and transcriptome data for the known ETT protein interactor REPLUMLESS (RPL). This analysis showed that among the 77 IAA-dependent targets of ETT, 24 were also bound and regulated by RPL, with 7 loci bound by both proteins in the same region of the gene. Detailed analysis of four of these genes supported a model in which the specificity in ETT regulation arises from its different partner proteins.
To demonstrate the usefulness of this set of auxin-responsive ETT direct target candidates, the authors explored the regulation of a few in more detail. In light of the previously proposed model that IAA prompts dissociation of ETT from a partner that mediates binding to a specific gene locus, they made use of available ChIP and transcriptome data for the known ETT protein interactor REPLUMLESS (RPL). This analysis showed that among the 77 IAA-dependent targets of ETT, 24 were also bound and regulated by RPL, with 7 loci bound by both proteins in the same region of the gene. Detailed analysis of four of these genes supported a model in which the specificity in ETT regulation arises from its different partner proteins.
In all, this impressive work provides a solid foundation from which to understand the multiple actions of this non-canonical auxin signaling protein. The findings indicate that ETT is a transcriptional regulator that functions both in response to and independently of auxin and can serve as a positive or negative regulator. The results also support the importance of ETT as a mediator of hormone responses in floral development and point to the need to identify more of ETT’s protein partners, which likely provide specificity in its regulation of targets.
REFERENCES
Simonini, S., Bencivenga, S., Trick, M. and Østergaard, L. (2017). Auxin-Induced Modulation of ETTIN Activity Orchestrates Gene Expression in Arabidopsis. Plant Cell. doi: 10.1105/tpc.17.00389.
Simonini, S., Deb, J., Moubayidin, L., Stephenson, P., Valluru, M., Freire-Rios, A., Sorefan, K., Weijers, D., Friml, J., and Østergaard, L. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30: 2286-2296.
Weijers, D., and Wagner, D. (2016). Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 67: 539-574.




Leave a Reply
Want to join the discussion?Feel free to contribute!