What We’re Reading: September 28
Focused Review: A role for ecophysiology in the ’omics’ era
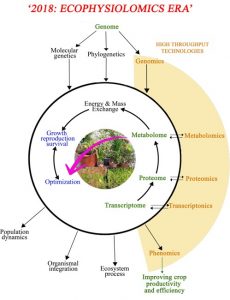 Ecophysiology is the study of plant functioning as modulated by the environment (or, as described by one author, “outdoors physiology“). Flexas and Gago ask whether research (and training) in ecophysiology has been left behind somewhat by successes in -omics approaches, and from a Web of Science query they conclude that ecophysiology research has been negatively affected by the rise in -omics. The authors provide two examples where ecophysiological and -omics strategies have come to opposite conclusions, and call for a greater intergration of the two approaches. They suggest that researchers could use more realistic growth conditions as starting points for -omics studies, and also collect ecophysiological measurements alongside -omics data. They also argue for the incorporation of more diverse plants into -omics studies, especially plants adapted to extreme environments that really challenge physiological systems. The authors conclude by highlighting the need “to integrate the new disciplines with pre‐existing ecophysiology theoretical knowledge and methodological skills to maximize the understanding of plants from a multidisciplinary approach,” and for “both communities to make steps to approach each other and collaborate.” (Summary by Mary Williams) Plant J. 10.1111/tpj.14059
Ecophysiology is the study of plant functioning as modulated by the environment (or, as described by one author, “outdoors physiology“). Flexas and Gago ask whether research (and training) in ecophysiology has been left behind somewhat by successes in -omics approaches, and from a Web of Science query they conclude that ecophysiology research has been negatively affected by the rise in -omics. The authors provide two examples where ecophysiological and -omics strategies have come to opposite conclusions, and call for a greater intergration of the two approaches. They suggest that researchers could use more realistic growth conditions as starting points for -omics studies, and also collect ecophysiological measurements alongside -omics data. They also argue for the incorporation of more diverse plants into -omics studies, especially plants adapted to extreme environments that really challenge physiological systems. The authors conclude by highlighting the need “to integrate the new disciplines with pre‐existing ecophysiology theoretical knowledge and methodological skills to maximize the understanding of plants from a multidisciplinary approach,” and for “both communities to make steps to approach each other and collaborate.” (Summary by Mary Williams) Plant J. 10.1111/tpj.14059
Opinion: Best practice data life cycle approaches for the life sciences
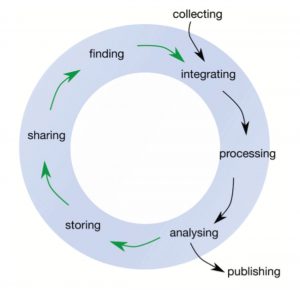 Producing data is costly, and with advances in technology, we are producing data with revolutionary capacity. However, we are also losing data at an astonishing rate, with around 80% of data being unavailable 20 years after publication. Griffin et al. provide guidelines, repositories and other resources that can be used by scientists to deposit and document their data. The reasons to deposit data and code in open source repositories are altruistic as well as selfish, as data shared according to FAIR principles boost citation indices and ensure eligibility for funding. The authors stress the importance of including training on effective data sharing in the curriculum, ensuring data are FAIR in the future and extending data lifespans. (Summary by Magdalena Julkowska) F1000 Res. 10.12688/f1000research.12344.2
Producing data is costly, and with advances in technology, we are producing data with revolutionary capacity. However, we are also losing data at an astonishing rate, with around 80% of data being unavailable 20 years after publication. Griffin et al. provide guidelines, repositories and other resources that can be used by scientists to deposit and document their data. The reasons to deposit data and code in open source repositories are altruistic as well as selfish, as data shared according to FAIR principles boost citation indices and ensure eligibility for funding. The authors stress the importance of including training on effective data sharing in the curriculum, ensuring data are FAIR in the future and extending data lifespans. (Summary by Magdalena Julkowska) F1000 Res. 10.12688/f1000research.12344.2
Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa
 The recent focus on quinoa is a result of not only its high nutritional value, but also its astonishing salt stress tolerance. Epidermal bladder cells (EBC) are suspected to be a cause of salinity tolerance by serving as salt dumping place. In order to identify molecular mechanisms underlying salt sequestration, Bohm et al. performed transcriptome analysis of EBC under control and salt stress conditions. The salt-induced changes in the EBC transcriptome were minimal. The group identified a number of transporters specific to EBC involved in sodium loading into the EBC (CqHKT1.2) and chloride transport into the vacuole of EBC (CqClc-c). Although the leaf was identified as the major proline production site, EBC-specific potassium and proline transporters were hypothesized to keep the EBC osmotically balanced. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.08.004
The recent focus on quinoa is a result of not only its high nutritional value, but also its astonishing salt stress tolerance. Epidermal bladder cells (EBC) are suspected to be a cause of salinity tolerance by serving as salt dumping place. In order to identify molecular mechanisms underlying salt sequestration, Bohm et al. performed transcriptome analysis of EBC under control and salt stress conditions. The salt-induced changes in the EBC transcriptome were minimal. The group identified a number of transporters specific to EBC involved in sodium loading into the EBC (CqHKT1.2) and chloride transport into the vacuole of EBC (CqClc-c). Although the leaf was identified as the major proline production site, EBC-specific potassium and proline transporters were hypothesized to keep the EBC osmotically balanced. (Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.08.004
SHOU4 proteins regulate trafficking of cellulose synthase complexes to the plasma membrane
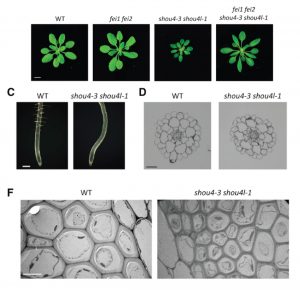 Plant cell walls provide mechanical support and define the extent and direction of cell expansion. Regulation of cellulose synthase determines cell wall load bearing and involves the receptor like kinases FEI1/FEI2. To gain further understanding of cell wall integrity regulation Polko et al. performed a suppressor screen of fei1/fei2. The group identified SHOU4, encoding a novel transmembrane protein showing no homology to previously identified functional domains. SHOU4 localized at the plasma membrane, where it acted as a negative regulator of cellulose synthase exocytosis. Functional mutation of SHOU4 and its closest paralog, SHOU4-like, resulted in dwarfed growth, thinner cell walls and aborted root hair development. Those results illustrate importance of mechanisms involved in cellulose synthase delivery to plasma membrane in developmental processes. Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.07.076
Plant cell walls provide mechanical support and define the extent and direction of cell expansion. Regulation of cellulose synthase determines cell wall load bearing and involves the receptor like kinases FEI1/FEI2. To gain further understanding of cell wall integrity regulation Polko et al. performed a suppressor screen of fei1/fei2. The group identified SHOU4, encoding a novel transmembrane protein showing no homology to previously identified functional domains. SHOU4 localized at the plasma membrane, where it acted as a negative regulator of cellulose synthase exocytosis. Functional mutation of SHOU4 and its closest paralog, SHOU4-like, resulted in dwarfed growth, thinner cell walls and aborted root hair development. Those results illustrate importance of mechanisms involved in cellulose synthase delivery to plasma membrane in developmental processes. Summary by Magdalena Julkowska) Curr. Biol. 10.1016/j.cub.2018.07.076
ZmbZIP22 regulates 27-kD γ-zein transcription during maize endosperm development
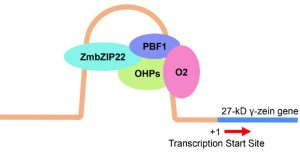 Zeins are maize seed storage proteins, but are notoriously low in the amino acids lycine and tryptophan. Therefore, to improve maize’s nutritional properties, efforts have been made to decrease zein levels. However, such efforts often lead to abnormal (opague) kernels. Previous work has demonstrated that the 27-kD γ-zein gene is extremely highly expressed in the maize endosperm, and several transcription factors that are involved in its expression have been identified. However, even when all of these are mutated, the gene is still expressed, suggesting that other transcription factors contribute. Li et al. set about identifying and characterizing this missing partner, ZmbZIP22. Using a combination of tranditional (gel shift) and modern (CRISPR/Cas9, RNA-seq, ChIP-seq) methods, the identified cis-elements bound by ZmbZIP22 and genes it is involved in regulating. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00422
Zeins are maize seed storage proteins, but are notoriously low in the amino acids lycine and tryptophan. Therefore, to improve maize’s nutritional properties, efforts have been made to decrease zein levels. However, such efforts often lead to abnormal (opague) kernels. Previous work has demonstrated that the 27-kD γ-zein gene is extremely highly expressed in the maize endosperm, and several transcription factors that are involved in its expression have been identified. However, even when all of these are mutated, the gene is still expressed, suggesting that other transcription factors contribute. Li et al. set about identifying and characterizing this missing partner, ZmbZIP22. Using a combination of tranditional (gel shift) and modern (CRISPR/Cas9, RNA-seq, ChIP-seq) methods, the identified cis-elements bound by ZmbZIP22 and genes it is involved in regulating. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00422
Flowering time in the real world
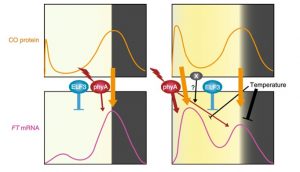 There are pros and cons to growing plants in controlled conditions. On the one hand, controlling light, temperature, humidity and other environmental factors should aid reproducibility between experiments and labs. But what if the conditions used profoundly and unexpectedly affect the process you are studying? This is what Song et al. show in their new work. They found that Arabidopsis plants grown outdoors in natural conditions flowered with fewer leaves than those in growth chambers (even under long-day conditions), and showed a correspondingly different expression pattern of the florigen FT gene. The authors were better able to mimic the natural growth and expression patterns by supplementing standard growth chamber conditions with additional far-red light, and introducing daily temperature fluctuations. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0253-3
There are pros and cons to growing plants in controlled conditions. On the one hand, controlling light, temperature, humidity and other environmental factors should aid reproducibility between experiments and labs. But what if the conditions used profoundly and unexpectedly affect the process you are studying? This is what Song et al. show in their new work. They found that Arabidopsis plants grown outdoors in natural conditions flowered with fewer leaves than those in growth chambers (even under long-day conditions), and showed a correspondingly different expression pattern of the florigen FT gene. The authors were better able to mimic the natural growth and expression patterns by supplementing standard growth chamber conditions with additional far-red light, and introducing daily temperature fluctuations. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0253-3