What we’re reading: October 13th
Update: Peroxisome function, biogenesis, and dynamics in plants
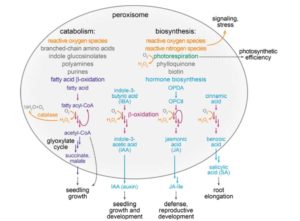 Peroxisomes are endoplasmic reticulum-derived membrane-enclosed organelles in which many oxidative enzymatic reactions are compartmentalized. These reactions and their products contribute to energy production, detoxification, and signaling. Kao et al. review our understanding of the plant peroxisome, including the role of peroxisome enzymes (and how these nuclear-encoded proteins are targeted to the peroxisome), as well as the biogenesis of peroxisomes themselves and the contributions of peroxins (PEX proteins). The process of peroxisome turnover (pexophagy, a form of autophagy) is also described. Plant Physiol. 10.1104/pp.17.01050
Peroxisomes are endoplasmic reticulum-derived membrane-enclosed organelles in which many oxidative enzymatic reactions are compartmentalized. These reactions and their products contribute to energy production, detoxification, and signaling. Kao et al. review our understanding of the plant peroxisome, including the role of peroxisome enzymes (and how these nuclear-encoded proteins are targeted to the peroxisome), as well as the biogenesis of peroxisomes themselves and the contributions of peroxins (PEX proteins). The process of peroxisome turnover (pexophagy, a form of autophagy) is also described. Plant Physiol. 10.1104/pp.17.01050
Update: Flower primary metabolism, pollinators’ preferences and seed and fruit set
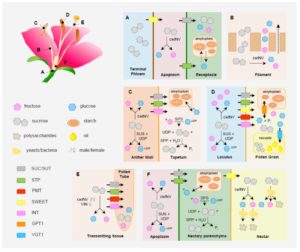 Because of their important roles in attracting pollinators, the secondary or specialized metabolites of flowers (color, fragrance) get a lot of attention. Borghi and Fernie argue that flower function is equally dependent on primary (central) metabolites, which not only provide the precursors for secondary metabolite production but also serve as food for pollinators and support the development of fruits and seeds. In their review, they discuss the metabolism, transport and partitioning of sugars and amino acids through floral organs. Challenges to fully elucidating these pathways include genetic redundancy, a tendency to overlook floral phenotypes in mutant studies, and and the common practice of treating flowers as a single unit, “despite the fact that in no other plant organ as much as in flowers, tissue compartmentalization underlies function.” The authors also describe the contributions of metabolites to pollen tube growth and the roles of microbes in altering nectar chemistry and thus pollinator behavior. Plant Physiol. 10.1104/pp.17.01164
Because of their important roles in attracting pollinators, the secondary or specialized metabolites of flowers (color, fragrance) get a lot of attention. Borghi and Fernie argue that flower function is equally dependent on primary (central) metabolites, which not only provide the precursors for secondary metabolite production but also serve as food for pollinators and support the development of fruits and seeds. In their review, they discuss the metabolism, transport and partitioning of sugars and amino acids through floral organs. Challenges to fully elucidating these pathways include genetic redundancy, a tendency to overlook floral phenotypes in mutant studies, and and the common practice of treating flowers as a single unit, “despite the fact that in no other plant organ as much as in flowers, tissue compartmentalization underlies function.” The authors also describe the contributions of metabolites to pollen tube growth and the roles of microbes in altering nectar chemistry and thus pollinator behavior. Plant Physiol. 10.1104/pp.17.01164
Insights into land plant evolution garnered from the Marchantia polymorpha genome
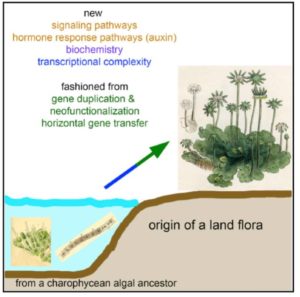 The liverwort Marchantia polymorpha is a fascinating plant for many reasons, including the fact that it is one of the earliest terrestrial species that split off from the rest of the land plants. Therefore, comparisons between Marchantia and green algae or Marchantia and the rest of the land plants tell us a lot about the characteristics of the earliest land plants. Bowman et al. present the first liverwort genome and share what it reveals about the evolution of land plants. This study provides a snapshot of the timing of the evolution of diverse plant processes, and spans phytohormone signaling, control of hydration, and defense responses. The authors also touch on the contribution of horizontal-gene transfer from fungi in supporting the colonization of land by early plants, and ask the question of why whole-genome duplications are lacking in Marchantia. Cell 10.1016/j.cell.2017.09.030
The liverwort Marchantia polymorpha is a fascinating plant for many reasons, including the fact that it is one of the earliest terrestrial species that split off from the rest of the land plants. Therefore, comparisons between Marchantia and green algae or Marchantia and the rest of the land plants tell us a lot about the characteristics of the earliest land plants. Bowman et al. present the first liverwort genome and share what it reveals about the evolution of land plants. This study provides a snapshot of the timing of the evolution of diverse plant processes, and spans phytohormone signaling, control of hydration, and defense responses. The authors also touch on the contribution of horizontal-gene transfer from fungi in supporting the colonization of land by early plants, and ask the question of why whole-genome duplications are lacking in Marchantia. Cell 10.1016/j.cell.2017.09.030
Insights into plasmodesmata composition of fully developed Arabidopsis thaliana leaves
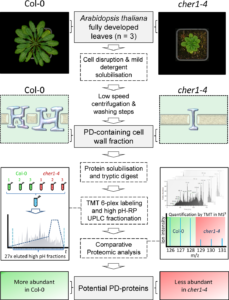 Plasmodesmata (PD) are complex, regulated channels between plant cells that facilitate the movement of signals, metabolites and pathogens, but their small size makes them difficult to study. Previously, Kraner et al. identified an Arabidopsis mutant that produced fewer, simpler plasmodesmata. In their new work, they used this mutant as a tool for a comparative proteomic study to identify proteins associated with plasmodesmata (i.e., proteins under-represented in the cell-wall fraction of the mutant). They identified “61 potential PD candidates containing 17 previously described PD-associated proteins”. This work lays the foundation for further research into the structure and function of plasmodesmata. Plant J. 10.1111/tpj.13702
Plasmodesmata (PD) are complex, regulated channels between plant cells that facilitate the movement of signals, metabolites and pathogens, but their small size makes them difficult to study. Previously, Kraner et al. identified an Arabidopsis mutant that produced fewer, simpler plasmodesmata. In their new work, they used this mutant as a tool for a comparative proteomic study to identify proteins associated with plasmodesmata (i.e., proteins under-represented in the cell-wall fraction of the mutant). They identified “61 potential PD candidates containing 17 previously described PD-associated proteins”. This work lays the foundation for further research into the structure and function of plasmodesmata. Plant J. 10.1111/tpj.13702
Pectinase function in growth and stomatal dynamics
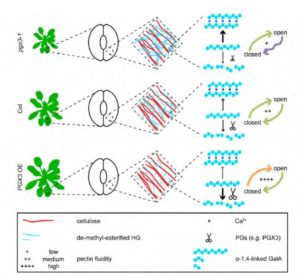 Pectin is a polymer that holds cell walls together and stiffens the walls, but what happens when those cell walls need to move, for example when cells (including guard cells) expand? Rui et al. started with a guard-cell specific transcriptome to identify genes encoding cell-wall modifying enzymes. They identified POLYGALACTURONASE INVOLVED IN EXPANSION3 (PGX3), a guard-cell enriched and ABA-inducible pectinase-encoding gene. The authors show that elevated PGX3 expression affects pectin molecular mass and abundance, thus affecting cell wall mechanics and leading to increased cell expansion and enhanced stomatal pore closure, whereas pgx3 mutants are delayed in stomatal closure. They present a model for how PGX3 affects cell wall stiffness and stomatal movements. Plant Cell 10.1105/tpc.17.00568
Pectin is a polymer that holds cell walls together and stiffens the walls, but what happens when those cell walls need to move, for example when cells (including guard cells) expand? Rui et al. started with a guard-cell specific transcriptome to identify genes encoding cell-wall modifying enzymes. They identified POLYGALACTURONASE INVOLVED IN EXPANSION3 (PGX3), a guard-cell enriched and ABA-inducible pectinase-encoding gene. The authors show that elevated PGX3 expression affects pectin molecular mass and abundance, thus affecting cell wall mechanics and leading to increased cell expansion and enhanced stomatal pore closure, whereas pgx3 mutants are delayed in stomatal closure. They present a model for how PGX3 affects cell wall stiffness and stomatal movements. Plant Cell 10.1105/tpc.17.00568
Ectopic expression of WINDING 1 leads to asymmetrical distribution of auxin and a spiral phenotype in rice
 Through insertional mutagenesis of rice, Cheng et al. identified a BTBN (Brac/Tramtrack/Broad complex and NPH3 domain)-encoding gene which they named WINDING 1 (WIN1), because the mutant phenotype (caused by ectopic expression in the shoot) causes the shoot to grow in a spiral. Ectopic expression of WIN1 in rice causes an asymmetric distribution of auxin and differential cell elongation in the shoot. The gene, which falls into a monocot-specific clade, has no effect when ectopically expressed in Arabidopsis. In rice, the WIN1 protein normally localizes to the plasma membrane and is enriched at plasmodesmata. The authors propose that WIN1 has a role in auxin redistribution through plasmodesmata. Plant Cell Physiol. 10.1093/pcp/pcx088
Through insertional mutagenesis of rice, Cheng et al. identified a BTBN (Brac/Tramtrack/Broad complex and NPH3 domain)-encoding gene which they named WINDING 1 (WIN1), because the mutant phenotype (caused by ectopic expression in the shoot) causes the shoot to grow in a spiral. Ectopic expression of WIN1 in rice causes an asymmetric distribution of auxin and differential cell elongation in the shoot. The gene, which falls into a monocot-specific clade, has no effect when ectopically expressed in Arabidopsis. In rice, the WIN1 protein normally localizes to the plasma membrane and is enriched at plasmodesmata. The authors propose that WIN1 has a role in auxin redistribution through plasmodesmata. Plant Cell Physiol. 10.1093/pcp/pcx088
In vivo gibberellin gradients visualized in rapidly elongating tissues
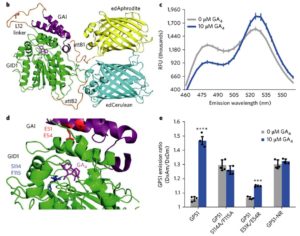 Biosensors can reveal cellular or even sub-cellular distributions of hormones, greatly enhancing our understanding of hormone action. Rizza et al. have developed an in vivo, FRET-based gibberellin biosensor, GIBBERELLIN PERCEPTION SENSOR 1 (GPS1), that incorporates portions of two GA-binding proteins, (the receptor GID1 and a DELLA protein that GID1 interacts with when bound to GA) as well as Förster Resonance Energy Transfer (FRET) donor and recipient modules. In the presence of GA, a conformational change in GPS1 facilitates energy transfer which is measurable by an increase in ratio of acceptor emission over donor emission. Using GPS1 the authors showed a correlation of endogenous and exogenous GA levels with cell elongation, although the contributions of synthesis, transport and catabolism to the formation of these distribution patterns remains uncertain. They also showed evidence for PIF transcription factors in promoting GA accumulation in the dark, suggesting a “PIF positive feedback loop through GA signaling”. Nature Plants 10.1038/s41477-017-0021-9.
Biosensors can reveal cellular or even sub-cellular distributions of hormones, greatly enhancing our understanding of hormone action. Rizza et al. have developed an in vivo, FRET-based gibberellin biosensor, GIBBERELLIN PERCEPTION SENSOR 1 (GPS1), that incorporates portions of two GA-binding proteins, (the receptor GID1 and a DELLA protein that GID1 interacts with when bound to GA) as well as Förster Resonance Energy Transfer (FRET) donor and recipient modules. In the presence of GA, a conformational change in GPS1 facilitates energy transfer which is measurable by an increase in ratio of acceptor emission over donor emission. Using GPS1 the authors showed a correlation of endogenous and exogenous GA levels with cell elongation, although the contributions of synthesis, transport and catabolism to the formation of these distribution patterns remains uncertain. They also showed evidence for PIF transcription factors in promoting GA accumulation in the dark, suggesting a “PIF positive feedback loop through GA signaling”. Nature Plants 10.1038/s41477-017-0021-9.
Genomic and epigenomic events occurring during germination
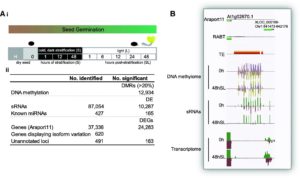 Seeds integrate environmental information and trigger molecular pathways that help with the decision making process of whether to germinate or not. Several studies in numerous species have shown details of global transcriptomic changes during germination. However, Narsai et al. dig a little deeper. By using next-generation sequencing, they explored not only transcriptomic changes (including unannotated loci), but also the occurrence of alternative splicing events, changes in small- and micro-RNAs expression, as well as changes in DNA methylation patterns. The paper is rich in data (publicly available) and certainly shows how dynamic are the molecular events that underlie seed germination. (Summary by Gaby Auge) Genome Biology 10.1186/s13059-017-1302-3
Seeds integrate environmental information and trigger molecular pathways that help with the decision making process of whether to germinate or not. Several studies in numerous species have shown details of global transcriptomic changes during germination. However, Narsai et al. dig a little deeper. By using next-generation sequencing, they explored not only transcriptomic changes (including unannotated loci), but also the occurrence of alternative splicing events, changes in small- and micro-RNAs expression, as well as changes in DNA methylation patterns. The paper is rich in data (publicly available) and certainly shows how dynamic are the molecular events that underlie seed germination. (Summary by Gaby Auge) Genome Biology 10.1186/s13059-017-1302-3
Temporal network analysis of mild drought in Brassica rapa
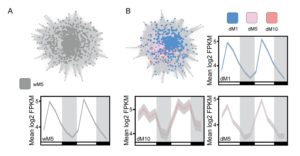 If you whithold water from a plant it eventually will wilt, but before this visible change there are other measurable effects and responses. However, many plant processes change cyclically over a 24-hour period independently of early drought responses, so it can be difficult to separate drought-responsive changes in physiology or gene expression against their dynamic baselines. Greenham, Guadagno et al. compared daily physiological (stomatal conductance, photosynthetic rate and Photosystem II efficiency) and gene expression (RNA-seq) levels in well-watered vs droughted Brassica rapa to identify those changes specific to the drought response. This then allowed them to correlate physiological responses to drought-specific transcriptional responses, “suggesting an early sensing of the drought treatment at the molecular level.” eLIFE 10.7554/eLife.29655.001
If you whithold water from a plant it eventually will wilt, but before this visible change there are other measurable effects and responses. However, many plant processes change cyclically over a 24-hour period independently of early drought responses, so it can be difficult to separate drought-responsive changes in physiology or gene expression against their dynamic baselines. Greenham, Guadagno et al. compared daily physiological (stomatal conductance, photosynthetic rate and Photosystem II efficiency) and gene expression (RNA-seq) levels in well-watered vs droughted Brassica rapa to identify those changes specific to the drought response. This then allowed them to correlate physiological responses to drought-specific transcriptional responses, “suggesting an early sensing of the drought treatment at the molecular level.” eLIFE 10.7554/eLife.29655.001
Phosphorus nutrition and root-associated fungal microbiota of nonmycorrhizal Arabis alpina
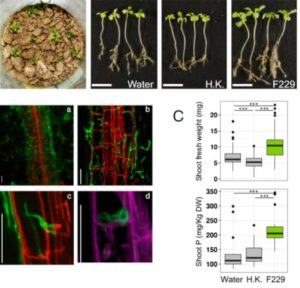 Associations with mycorrhizal fungi greatly enhance phosphorus (P) uptake for most plants, but the Brassicaceae are nonmycorrhizal due to the loss of essential symbiosis genes. Almario et al. investigated the fungal microbiota of Arabis alpina, a Brassicaceae species that grows in very-low P soils. The authors identified fungi of the Helotiales order that are strongly associated with roots in P-deficient conditions and which, when re-introduced into sterilized soil, enhanced P-uptake by the plants including the transfer of inorganic P from fungus to plant. As compared to related strains, this newly identified plant-beneficial endophyte shows an expansion of genes encoding carbohydrate-active enzymes (CAZymes), similar to that seen in mycorrhizal species. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1710455114
Associations with mycorrhizal fungi greatly enhance phosphorus (P) uptake for most plants, but the Brassicaceae are nonmycorrhizal due to the loss of essential symbiosis genes. Almario et al. investigated the fungal microbiota of Arabis alpina, a Brassicaceae species that grows in very-low P soils. The authors identified fungi of the Helotiales order that are strongly associated with roots in P-deficient conditions and which, when re-introduced into sterilized soil, enhanced P-uptake by the plants including the transfer of inorganic P from fungus to plant. As compared to related strains, this newly identified plant-beneficial endophyte shows an expansion of genes encoding carbohydrate-active enzymes (CAZymes), similar to that seen in mycorrhizal species. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1710455114
Low-gluten, non-transgenic wheat engineered with CRISPR/Cas9
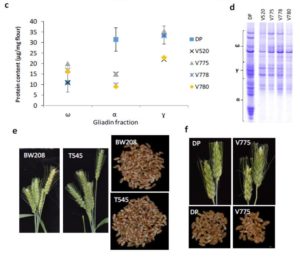 Celiac disease is a debilitating autoimmune disease in which antigens in plant gliadins (one type of gluten protein) stimulate production of antibodies that inflame the lining of the small intestine. In wheat, α-gliadin (the main cause of the sensitivity) is encoded by more than 100 genes, thwarting their elimination through traditional breeding methods. Sánchez-León et al. designed a CRISPR/Cas9 strategy to specifically induce mutations in α-gliadin-encoding genes. Amongst the lines examined, some showed a strong, heritable reduction in α-gliadin production. This study shows that potential application of CRISPR/Cas9 for the production of low-gluten wheat varieties. Plant Biotech. J. 10.1111/pbi.12837
Celiac disease is a debilitating autoimmune disease in which antigens in plant gliadins (one type of gluten protein) stimulate production of antibodies that inflame the lining of the small intestine. In wheat, α-gliadin (the main cause of the sensitivity) is encoded by more than 100 genes, thwarting their elimination through traditional breeding methods. Sánchez-León et al. designed a CRISPR/Cas9 strategy to specifically induce mutations in α-gliadin-encoding genes. Amongst the lines examined, some showed a strong, heritable reduction in α-gliadin production. This study shows that potential application of CRISPR/Cas9 for the production of low-gluten wheat varieties. Plant Biotech. J. 10.1111/pbi.12837
Peanuts that keep aflatoxin at bay: A threshold that matters
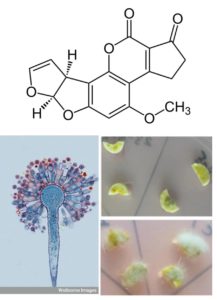 Aflatoxins are small molecules that are extremely damaging to human health that are produced by the fungal species Aspergillus flavus and Aspergillus parasiticus. Peanut pods form underground and so are particularly vulnerable to infection by the fungus. Sharma et al. used a two-pronged approach to develop peanuts that accumulate little or no aflatoxin. On the one hand, they introduced defensin genes from related legumes (Medicago sativa and Medicago truncatula; MsDef1 and MtDef4.2) that confer increased resistance to fungal infection. On the other hand, they introduced into the peanut host a construct that suppresses aflatoxin biosynthesis in the fungus through host-induced gene silencing (HIGS). Peanuts engineered with the defensin and HIGS contructs accumulated neglible amounts of aflatoxins. Plant Biotech. J. 10.1111/pbi.12846
Aflatoxins are small molecules that are extremely damaging to human health that are produced by the fungal species Aspergillus flavus and Aspergillus parasiticus. Peanut pods form underground and so are particularly vulnerable to infection by the fungus. Sharma et al. used a two-pronged approach to develop peanuts that accumulate little or no aflatoxin. On the one hand, they introduced defensin genes from related legumes (Medicago sativa and Medicago truncatula; MsDef1 and MtDef4.2) that confer increased resistance to fungal infection. On the other hand, they introduced into the peanut host a construct that suppresses aflatoxin biosynthesis in the fungus through host-induced gene silencing (HIGS). Peanuts engineered with the defensin and HIGS contructs accumulated neglible amounts of aflatoxins. Plant Biotech. J. 10.1111/pbi.12846