What We’re Reading: November 23rd
Editorial. Counting what counts: the importance of quantitative approaches to studying plant cell biology
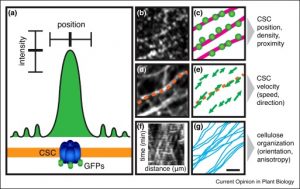 In the new Cell Biology issue of Current Opinion in Plant Biology, editors Haswell and Dixit have chosen to focus on quantitative cell biology, arguing that, “if seeing is believing, then measuring is knowing.” Topics of the issue’s reviews span parts of the cell (including cell wall, cytoskeleton, stromules), things cells do (including morphogenesis) and tools and models (including optical tweezers, Chlamydomonas, Physcomitrella, and guard cells). You can find all of the articles in this issue here. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.10.003
In the new Cell Biology issue of Current Opinion in Plant Biology, editors Haswell and Dixit have chosen to focus on quantitative cell biology, arguing that, “if seeing is believing, then measuring is knowing.” Topics of the issue’s reviews span parts of the cell (including cell wall, cytoskeleton, stromules), things cells do (including morphogenesis) and tools and models (including optical tweezers, Chlamydomonas, Physcomitrella, and guard cells). You can find all of the articles in this issue here. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.10.003
Review. Of mice and plants: Comparative developmental systems biology ($)
 Although mulicellularity, and so also development, evolved independently in animals and plants, there are nevertheless some striking parallels in the mechanisms used to coordinate these processes. Ten Tesschur compares four developmental processes in plants to similar processes that occur in animal development. As examples, root growth and lateral root prepatterning is compared to body axis elongation and somite segmentation, and plant shoot phyllotaxis is compared to digit patterning in a vertebrate limb. Through these comparisons, the author observes that general design principles and core mechanistic properties are conserved across developmental programs. She summarizes, “Recovering such design principles even in an across-kingdom comparison further underlines the fact that evolution of developmental processes is far from random but instead heavily biased by a limited number of mechanism existing for each particular patterning demand.” (Summary by Mary Williams) Devel. Biol. 10.1016/j.ydbio.2018.10.024
Although mulicellularity, and so also development, evolved independently in animals and plants, there are nevertheless some striking parallels in the mechanisms used to coordinate these processes. Ten Tesschur compares four developmental processes in plants to similar processes that occur in animal development. As examples, root growth and lateral root prepatterning is compared to body axis elongation and somite segmentation, and plant shoot phyllotaxis is compared to digit patterning in a vertebrate limb. Through these comparisons, the author observes that general design principles and core mechanistic properties are conserved across developmental programs. She summarizes, “Recovering such design principles even in an across-kingdom comparison further underlines the fact that evolution of developmental processes is far from random but instead heavily biased by a limited number of mechanism existing for each particular patterning demand.” (Summary by Mary Williams) Devel. Biol. 10.1016/j.ydbio.2018.10.024
Review: Perspective on wheat yield and quality with reduced nitrogen supply
 Wheat production demands huge inputs of nitrogen as fertilizer, with accompanying financial and environmental costs. Zorb et al. discuss several strategies by which to maintain wheat yields and wheat quality while decreasing N use and wastage. Some of these are agronomic, such as ensuring that the N applications occur at times that are optimal for uptake and mobilization to the grain; N assimilation and reallocation efficiencies differ with developmental stage. Other approaches involve genetic strategies, focusing on varieties that maintain grain qualities even with less N application. The authors observe that of the different proteins that accumulate in the grain, only some have a role in the bread dough qualites of elasticity and viscosity. At least for animal food, it should be possible to decrease N inputs by eliminating or lowering the amounts of these non-bread-related proteins. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.08.012
Wheat production demands huge inputs of nitrogen as fertilizer, with accompanying financial and environmental costs. Zorb et al. discuss several strategies by which to maintain wheat yields and wheat quality while decreasing N use and wastage. Some of these are agronomic, such as ensuring that the N applications occur at times that are optimal for uptake and mobilization to the grain; N assimilation and reallocation efficiencies differ with developmental stage. Other approaches involve genetic strategies, focusing on varieties that maintain grain qualities even with less N application. The authors observe that of the different proteins that accumulate in the grain, only some have a role in the bread dough qualites of elasticity and viscosity. At least for animal food, it should be possible to decrease N inputs by eliminating or lowering the amounts of these non-bread-related proteins. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.08.012
Finding candidate genes in maize with Camoco
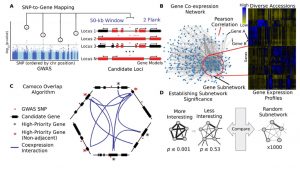 Schaefer and colleagues have developed Camoco (Co-analysis of molecular components), which integrates data from GWAS and co-expression networks to identify high confidence candidate genes associated with a phenotype of interest. To evaluate the program they used GWAS and co-expression data from maize. They used gene expression data from three datasets: data from whole-seedling transcriptome from diverse maize inbred lines, data from different maize tissues and data points from maize reference accession B73, and a dataset from roots from maize inbreds with different primary elemental absorption phenotypes. In the three cases the gene expression data came from RNA-seq, but microarray data could also be used. The evaluation showed that expression profiles from genetically diverse individuals produced more powerful co-expression networks compared to data from a single reference genotype. Two genes identify by Comoco in the root dataset were validated using mutants. The authors suggested that the identification of the best co-expression context (i.e. tissue) for a determined GWAS improves the odds of finding the genes associated to the phenotype of interest. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.18.00299
Schaefer and colleagues have developed Camoco (Co-analysis of molecular components), which integrates data from GWAS and co-expression networks to identify high confidence candidate genes associated with a phenotype of interest. To evaluate the program they used GWAS and co-expression data from maize. They used gene expression data from three datasets: data from whole-seedling transcriptome from diverse maize inbred lines, data from different maize tissues and data points from maize reference accession B73, and a dataset from roots from maize inbreds with different primary elemental absorption phenotypes. In the three cases the gene expression data came from RNA-seq, but microarray data could also be used. The evaluation showed that expression profiles from genetically diverse individuals produced more powerful co-expression networks compared to data from a single reference genotype. Two genes identify by Comoco in the root dataset were validated using mutants. The authors suggested that the identification of the best co-expression context (i.e. tissue) for a determined GWAS improves the odds of finding the genes associated to the phenotype of interest. (Summary by Cecilia Vasquez-Robinet) Plant Cell 10.1105/tpc.18.00299
POLAR-guided signalling complex assembly and localization drive asymmetric cell division
 In Arabidopsis, stomatal guard cells are specified following a set of conserved asymmetric cell divisions (ACDs), starting with a meristemoid mother cell. How do cells establish asymmetric divisions? Several proteins have been identified that are required for this, including BASL and POLAR, transcription factors including SPCH, as well as BIN2 and MAP kinases. BIN2 has been previously shown to phosphorylate both MAP kinases and SPCH. Houbaert et al. show that in the meristemoid mother cell, POLAR acts as a scaffold that is needed to polarize BIN2, which is necessary for assymetric divisions. When POLAR levels are low, as in the stomtatal lineage ground cell, BIN2 localizes to the nucleus where it phosphorylates SPCH and restricts ACDs. (Summary by Mary Williams) Nature 10.1038/s41586-018-0714-x
In Arabidopsis, stomatal guard cells are specified following a set of conserved asymmetric cell divisions (ACDs), starting with a meristemoid mother cell. How do cells establish asymmetric divisions? Several proteins have been identified that are required for this, including BASL and POLAR, transcription factors including SPCH, as well as BIN2 and MAP kinases. BIN2 has been previously shown to phosphorylate both MAP kinases and SPCH. Houbaert et al. show that in the meristemoid mother cell, POLAR acts as a scaffold that is needed to polarize BIN2, which is necessary for assymetric divisions. When POLAR levels are low, as in the stomtatal lineage ground cell, BIN2 localizes to the nucleus where it phosphorylates SPCH and restricts ACDs. (Summary by Mary Williams) Nature 10.1038/s41586-018-0714-x
Archetypal roles of an ABA receptor in drought and sugar responses in liverworts ($)
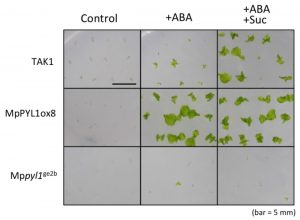 Much of our understanding of the molecular pathway for ABA response comes from studies on Arabidopsis. Thanks to genomic sequencing efforts, we know that this pathway is largely conserved amongst plants, including mosses. Jahan et al. extend this understanding to liverworts, drawing on the recently sequenced genome of Marchantia polymorpha. They identified five proteins as showing some homology to the PYL/PYR/RCAR ABA receptors, but found that only one, MpPYL1, showed all of the characteristics of these receptor proteins. They showed that overexpression of MpPYL1 led to greater ABA sensitivity, and knock-out mutations rendered the plants ABA insensitive. They also show that, like in angiosperms, ABA upregulates the LEA gene family, and mediates responses to exogenous sugars. These data support an ancestral role for the ABA receptors such as MpPYL1 in vegetative dessication tolerance in liverworts. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00761
Much of our understanding of the molecular pathway for ABA response comes from studies on Arabidopsis. Thanks to genomic sequencing efforts, we know that this pathway is largely conserved amongst plants, including mosses. Jahan et al. extend this understanding to liverworts, drawing on the recently sequenced genome of Marchantia polymorpha. They identified five proteins as showing some homology to the PYL/PYR/RCAR ABA receptors, but found that only one, MpPYL1, showed all of the characteristics of these receptor proteins. They showed that overexpression of MpPYL1 led to greater ABA sensitivity, and knock-out mutations rendered the plants ABA insensitive. They also show that, like in angiosperms, ABA upregulates the LEA gene family, and mediates responses to exogenous sugars. These data support an ancestral role for the ABA receptors such as MpPYL1 in vegetative dessication tolerance in liverworts. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00761
Genetic dissection of Fe-dependent signaling in root developmental responses to phosphate deficiency ($)
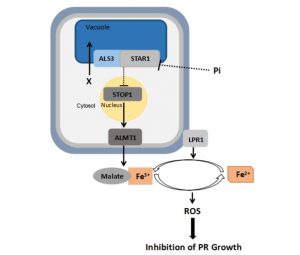 Phosphate deficiency leads to arrest of primary root elongation. Previous work has shown that this arrest depends upon Fe, raising the possibility that Fe accumulation in the root apex is the cause of growth arrest. Several genes involved in low Pi response have been identified, including genes encoding the LPR1 ferroxidase, ALS3/STAR1 transporter complex and the STOP-ALMT1 regulatory module; however, how these components interact has been unclear. Wang et al. carry out side-by-side comparisons of growth and Fe-uptake responses to Pi deficiency in various combinatorial mutants and overexpressor lines, allowing them to be placed into a genetic pathway of function, and revealing that “the degree of the inhibition of PR growth is not simply linked to the level of Fe accumulated in the RAM or elongation zone.” (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00907
Phosphate deficiency leads to arrest of primary root elongation. Previous work has shown that this arrest depends upon Fe, raising the possibility that Fe accumulation in the root apex is the cause of growth arrest. Several genes involved in low Pi response have been identified, including genes encoding the LPR1 ferroxidase, ALS3/STAR1 transporter complex and the STOP-ALMT1 regulatory module; however, how these components interact has been unclear. Wang et al. carry out side-by-side comparisons of growth and Fe-uptake responses to Pi deficiency in various combinatorial mutants and overexpressor lines, allowing them to be placed into a genetic pathway of function, and revealing that “the degree of the inhibition of PR growth is not simply linked to the level of Fe accumulated in the RAM or elongation zone.” (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00907
Plant extracellular vesicles contain diverse small RNA species and are enriched in 10-17 nucleotide “tiny” RNAs
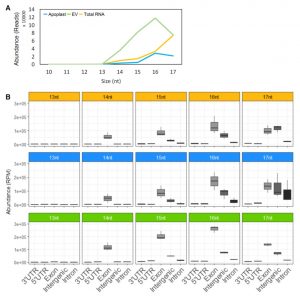 Small RNAs are known to move between cells and even between organisms. Studies in mammals have shown that extracellular vesicles (EVs) can contribute to small RNA transport, and recently small-RNA containing EVs were identified in plants. Baldrich et al. extend this work with an analysis of the types of small RNAs contained in plant EVs. Interestingly, while the typical distribution of small RNAs is dominated by 21-24 nt lengths, small RNAs in EVs are notably smaller than with the majority 10-17 nt, so are described as tiny RNAs (tyRNAs). Many of these appear to be derived from the loop region of processed miRNAs, in contrast to tyRNAs found outside of EVs, suggesting that the EVs are selectively packaged with a subset of tyRNAs. As yet, the function of these EVs and they tyRNAs they contain remain uncertain. (Summary by Mary Williams) bioRxiv 10.1101/472928
Small RNAs are known to move between cells and even between organisms. Studies in mammals have shown that extracellular vesicles (EVs) can contribute to small RNA transport, and recently small-RNA containing EVs were identified in plants. Baldrich et al. extend this work with an analysis of the types of small RNAs contained in plant EVs. Interestingly, while the typical distribution of small RNAs is dominated by 21-24 nt lengths, small RNAs in EVs are notably smaller than with the majority 10-17 nt, so are described as tiny RNAs (tyRNAs). Many of these appear to be derived from the loop region of processed miRNAs, in contrast to tyRNAs found outside of EVs, suggesting that the EVs are selectively packaged with a subset of tyRNAs. As yet, the function of these EVs and they tyRNAs they contain remain uncertain. (Summary by Mary Williams) bioRxiv 10.1101/472928
Brassica clubroot disease protist pathogen promotes phloem differentiation and expression of SWEET sucrose transporters
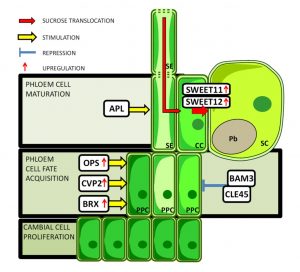 Clubroot is a disease of brassicas (including oilseed crops and Arabidopsis) caused by the protist Plasmodiophora brassicae. The infection is characterized by the formation of swollen tissues called galls that are metabolic sinks in which the pathogen propagates. Walerowski et al. took advantage of Arabidopsis tools to dissect the molecular events initiated by the pathogen. They show that pathogen infection induces genes that promote phloem differentiation including BREVIS RADIX (BRiX), COTYLEDON VASCULAR PATTERN 2 (CVP2) and OCTOPUS (OPS). Additionally, expression of the SWEET sucrose transporters increases. Collectively these changes contribute to increased flow of carbohydrates to the galls. Mutations that inhibit this metabolic hijacking inhibit gall formation and pathogen success. The authors conclude, “This work highlights the importance of precise regulation of sugar partitioning for plant-pathogen interactions and the dependence of P. brassicae’s performance on its capacity to induce a phloem sink at the feeding site.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00283
Clubroot is a disease of brassicas (including oilseed crops and Arabidopsis) caused by the protist Plasmodiophora brassicae. The infection is characterized by the formation of swollen tissues called galls that are metabolic sinks in which the pathogen propagates. Walerowski et al. took advantage of Arabidopsis tools to dissect the molecular events initiated by the pathogen. They show that pathogen infection induces genes that promote phloem differentiation including BREVIS RADIX (BRiX), COTYLEDON VASCULAR PATTERN 2 (CVP2) and OCTOPUS (OPS). Additionally, expression of the SWEET sucrose transporters increases. Collectively these changes contribute to increased flow of carbohydrates to the galls. Mutations that inhibit this metabolic hijacking inhibit gall formation and pathogen success. The authors conclude, “This work highlights the importance of precise regulation of sugar partitioning for plant-pathogen interactions and the dependence of P. brassicae’s performance on its capacity to induce a phloem sink at the feeding site.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00283



