What We’re Reading: May 18th
Review: Same tune, different song — cytokinins as virulence factors in plant–pathogen interactions?
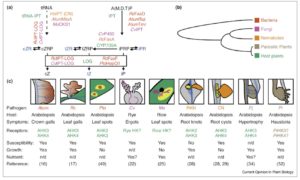 Many pathogens produce virulence factors that improve their pathogenicity, including in some cases compounds produced by the host, such as the hormone cytokinin. Spallek et al. review the various plant pathogens — spanning from bacteria to parasitic plants — that use cytokinins as virulence factors; cytokinin biosynthetic and modification genes can be found in these diverse pathogens. Cytokinin production can support pathogen growth through the development of structures such as tumors or galls or by altering patterns of nutrient transport, but can also enhance the host’s susceptibility to invasion. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.03.002
Many pathogens produce virulence factors that improve their pathogenicity, including in some cases compounds produced by the host, such as the hormone cytokinin. Spallek et al. review the various plant pathogens — spanning from bacteria to parasitic plants — that use cytokinins as virulence factors; cytokinin biosynthetic and modification genes can be found in these diverse pathogens. Cytokinin production can support pathogen growth through the development of structures such as tumors or galls or by altering patterns of nutrient transport, but can also enhance the host’s susceptibility to invasion. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.03.002
Review: The spring of systems biology-driven breeding ($)
 In this excellent review, Lavarenne et al. provide an accessible introduction to the tools and objectives of gene regulatory network (GRN) analysis. They include a glossary, show diagrams displaying different stages of GRN analysis, and provide links to key papers in which different methods have been applied to plant science. This article is recommended reading for anyone who is teaching or just wants to learn more about systems biology. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.04.005
In this excellent review, Lavarenne et al. provide an accessible introduction to the tools and objectives of gene regulatory network (GRN) analysis. They include a glossary, show diagrams displaying different stages of GRN analysis, and provide links to key papers in which different methods have been applied to plant science. This article is recommended reading for anyone who is teaching or just wants to learn more about systems biology. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.04.005
Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle ($)
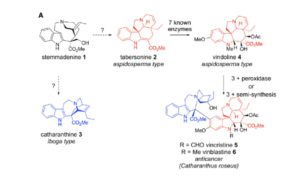 Plants make some amazing biologically active natural products, many with impressive applications such as vinblastine, a product of Catharantus roseus (Madagascar periwinkle). Caputi et al. identified key enzymes in vinblastine synthesis, opening the door for its production in alternative systems and the production of derivative compounds. It’s an interesting story of false-starts, unstable intermediates and hard-to-express enzymes, but it ends happily with a newly defined pathway and allows “the prospect of heterologous production of these expensive and valuable compounds in alternative host organisms.” (Summary by Mary Williams) Science 10.1126/science.aat4100
Plants make some amazing biologically active natural products, many with impressive applications such as vinblastine, a product of Catharantus roseus (Madagascar periwinkle). Caputi et al. identified key enzymes in vinblastine synthesis, opening the door for its production in alternative systems and the production of derivative compounds. It’s an interesting story of false-starts, unstable intermediates and hard-to-express enzymes, but it ends happily with a newly defined pathway and allows “the prospect of heterologous production of these expensive and valuable compounds in alternative host organisms.” (Summary by Mary Williams) Science 10.1126/science.aat4100
Stay CLASSY: Control of locus-specific de novo DNA methylation by the CLASSY family ($)
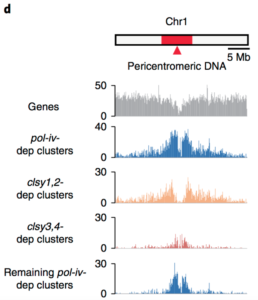 DNA methylation is fundamental for genome function and stability, including regulation of gene expression, silencing of transposable elements and control of recombination. While the processes involved in maintenance of DNA methylation are generally well understood, the factors required for locus-specific de novo establishment of DNA methylation remain unclear. In this study, Zhou et al. tested the role of chromatin remodelling factors from the CLASSY (CLSY) family in the de novo DNA methylation pathway in Arabidopsis. They showed that CLSYs interact with RNA polymerase IV and together mediate transcriptional silencing by controlling the production of 24-nt small-interfering RNA, which drive locus-specific de novo methylation. They further show that recruitment of CLSYs is driven by local enrichment in repressive chromatin modifications, such as DNA methylation in CG context and H3K9me2. Together, this study provides significant advances in understanding the initiation of de novo DNA methylation and highlights the interdependence between chromatin-based processes. (Summary by Matthias Benoit) Nature Genet. 10.1038/s41588-018-0115-y
DNA methylation is fundamental for genome function and stability, including regulation of gene expression, silencing of transposable elements and control of recombination. While the processes involved in maintenance of DNA methylation are generally well understood, the factors required for locus-specific de novo establishment of DNA methylation remain unclear. In this study, Zhou et al. tested the role of chromatin remodelling factors from the CLASSY (CLSY) family in the de novo DNA methylation pathway in Arabidopsis. They showed that CLSYs interact with RNA polymerase IV and together mediate transcriptional silencing by controlling the production of 24-nt small-interfering RNA, which drive locus-specific de novo methylation. They further show that recruitment of CLSYs is driven by local enrichment in repressive chromatin modifications, such as DNA methylation in CG context and H3K9me2. Together, this study provides significant advances in understanding the initiation of de novo DNA methylation and highlights the interdependence between chromatin-based processes. (Summary by Matthias Benoit) Nature Genet. 10.1038/s41588-018-0115-y
Integration of two RAB5 groups during endosomal transport in plants (OA)
 RAB proteins are membrane-anchored proteins that coordinate and regulate vesicle trafficking. Plants contain two group of RAB5 proteins, canonical (ARA7, RHA1) and plant-specific (ARA6), which share a common activator, VPS9A (Vacuolar Protein Sorting 9A). Ito et al. used an activated (GTP-bound) version of the plant-specific ARA6 to find interacting partner proteins. From that screening, they identified PUF2 (PLANT UNIQUE RAB5 EFFECTOR 2) as an effector molecule of ARA6. Surprisingly, PUF2 also interacts with inactivated (GDP-bound) canonical RAB5 and the common RAB5 effector (VPS9A). Based on biochemical and genetic evidence, the authors found that PUF2 recruits VPS9A as well as inactive canonical RAB5 to the endosomal membrane and facilitates its activation. However, ARA6 negatively regulates the PUF2-mediated RAB5 activation. This mechanistic model explained the pivotal role of PUF2 to regulate and integrate two groups of RAB5 in plant. (Summary by Arif Ashraf) eLife: 10.7554/eLife.34064.001
RAB proteins are membrane-anchored proteins that coordinate and regulate vesicle trafficking. Plants contain two group of RAB5 proteins, canonical (ARA7, RHA1) and plant-specific (ARA6), which share a common activator, VPS9A (Vacuolar Protein Sorting 9A). Ito et al. used an activated (GTP-bound) version of the plant-specific ARA6 to find interacting partner proteins. From that screening, they identified PUF2 (PLANT UNIQUE RAB5 EFFECTOR 2) as an effector molecule of ARA6. Surprisingly, PUF2 also interacts with inactivated (GDP-bound) canonical RAB5 and the common RAB5 effector (VPS9A). Based on biochemical and genetic evidence, the authors found that PUF2 recruits VPS9A as well as inactive canonical RAB5 to the endosomal membrane and facilitates its activation. However, ARA6 negatively regulates the PUF2-mediated RAB5 activation. This mechanistic model explained the pivotal role of PUF2 to regulate and integrate two groups of RAB5 in plant. (Summary by Arif Ashraf) eLife: 10.7554/eLife.34064.001
Synthetic hormone-responsive transcription factors can monitor and re-program plant development
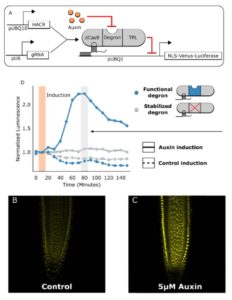 Khakhar et al. have developed a system by which genes can be made exquisitely responsive to specific hormones. They take advantage of the specificity of the guide DNA/Cas9 system, but use a deactivated (dCas9) enzyme that targets but does not cleave the target DNA. By fusing dCAS9 to a hormone-responsive degron (protein degradation domain) and a portion of the TOPLESS transcriptional repressor, they generated “hormone-activated Cas9-based repressors (HACRs, pronounced ‘hackers’).” The presence of the hormone (auxin, jasmonates or gibberellins) leads to degradation of the synthetic protein, enabling transcription. Using fluorescent proteins, they demonstrate patterns of endogenous protein accumulation, which correspond to data obtained through other methods. By targeting the HACR against genes encoding PIN1 auxin transporters, they are able to modulate plant development in predictable ways. (Summary by Mary Williams) eLIFE 10.7554/eLife.34702
Khakhar et al. have developed a system by which genes can be made exquisitely responsive to specific hormones. They take advantage of the specificity of the guide DNA/Cas9 system, but use a deactivated (dCas9) enzyme that targets but does not cleave the target DNA. By fusing dCAS9 to a hormone-responsive degron (protein degradation domain) and a portion of the TOPLESS transcriptional repressor, they generated “hormone-activated Cas9-based repressors (HACRs, pronounced ‘hackers’).” The presence of the hormone (auxin, jasmonates or gibberellins) leads to degradation of the synthetic protein, enabling transcription. Using fluorescent proteins, they demonstrate patterns of endogenous protein accumulation, which correspond to data obtained through other methods. By targeting the HACR against genes encoding PIN1 auxin transporters, they are able to modulate plant development in predictable ways. (Summary by Mary Williams) eLIFE 10.7554/eLife.34702
Regulation of Arabidopsis hypocotyl growth by transcriptional regulators
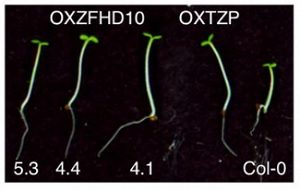 The integration of light-based environmental signals with photoreceptors and transcriptional regulators is essential for coordinating plant growth, development and energy production. One such integrator of light and signaling components is the Arabidopsis thaliana TANDEM ZINC-FINGER PLUS3 (TZP). TZP is a positive regulator of hypocotyl elongation and interacts with ZINC-FINGER HOMEODOMAIN 10 (ZFHD10). Perrella and colleagues use genomic studies, mutant studies and transcriptional analyses to characterize the function of TZP and ZFHD10 in the regulation of blue light-dependent transcription factors. ZFHD10 recruits TZP to promoters of key light-regulated genes. In conclusion, in response to blue light, ZFHD10 acts as a positive regulator of hypocotyl growth through the action of light signaling components. (Summary by Julia Miller) Proc. Natl. Acad. Sci. USA
The integration of light-based environmental signals with photoreceptors and transcriptional regulators is essential for coordinating plant growth, development and energy production. One such integrator of light and signaling components is the Arabidopsis thaliana TANDEM ZINC-FINGER PLUS3 (TZP). TZP is a positive regulator of hypocotyl elongation and interacts with ZINC-FINGER HOMEODOMAIN 10 (ZFHD10). Perrella and colleagues use genomic studies, mutant studies and transcriptional analyses to characterize the function of TZP and ZFHD10 in the regulation of blue light-dependent transcription factors. ZFHD10 recruits TZP to promoters of key light-regulated genes. In conclusion, in response to blue light, ZFHD10 acts as a positive regulator of hypocotyl growth through the action of light signaling components. (Summary by Julia Miller) Proc. Natl. Acad. Sci. USA
Sulfur partitioning between glutathione and protein synthesis determines plant growth
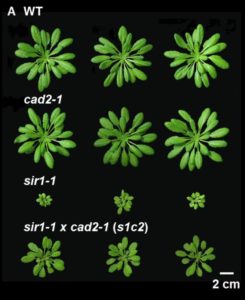 Sulfur is incorporated into the amino acid cysteine (Cys) and also the reactive-oxygen scavenger glutathione (GSH). Speiser et al. investigated the effect of restricting both Cys and glutathione production through a double-mutant analysis in Arabidopsis; sir1-1 is a slow-growing sulfite-reductase deficient mutant with limited production of Cys production, and cad2-1 is cadmium sensitive due to a restriction in glutathione production. Interestingly, the sir1-1 cad2-1 double mutant is significantly larger than the sir1-1 single mutant. Further analysis indicated that there is an increased flux through the assimilatory sulfate reduction pathway in the double mutant. Furthermore, TOR (Target of Rapamycin) activity is induced in the cad2-1 mutant; TOR is a metabolic sensor kinase and a regulator of protein synthesis and growth in all eukaryotes. This study provides new evidence for the role of TOR in regulating plant growth. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00421
Sulfur is incorporated into the amino acid cysteine (Cys) and also the reactive-oxygen scavenger glutathione (GSH). Speiser et al. investigated the effect of restricting both Cys and glutathione production through a double-mutant analysis in Arabidopsis; sir1-1 is a slow-growing sulfite-reductase deficient mutant with limited production of Cys production, and cad2-1 is cadmium sensitive due to a restriction in glutathione production. Interestingly, the sir1-1 cad2-1 double mutant is significantly larger than the sir1-1 single mutant. Further analysis indicated that there is an increased flux through the assimilatory sulfate reduction pathway in the double mutant. Furthermore, TOR (Target of Rapamycin) activity is induced in the cad2-1 mutant; TOR is a metabolic sensor kinase and a regulator of protein synthesis and growth in all eukaryotes. This study provides new evidence for the role of TOR in regulating plant growth. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.00421
A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling
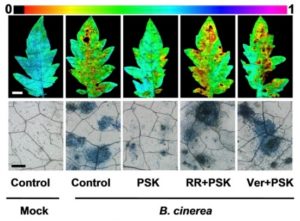 Plants are exposed to a wide variety of biotic stressors such as herbivores and microbial pathogens. Thus, they have developed a sophisticated response system which involves different signal molecules and accurate signaling pathway responses. Among them, phytosulfokine (PSK), a disulfated pentapeptide, has been suggested to be a key signaling molecule, but evidence that supports this fact has been scarce. In this report, Zhang et al. show that PSK is a damage-associated signaling molecule with potential to be used to enhance disease resistance in different plant species. They show that PSK biosynthesis, modification and perception are very important for plant immunity related to tomato responses against the fungal pathogen Botrytis cinerea, and that PSK activation of the PSK receptor 1, RPSK1, enhances calcium cytosolic concentration. This increase in calcium content is sensed by the calcium binding protein calmodulin which then interacts with YUCs (auxin biosynthesis enzymes) initiating auxin-dependent immune responses. (Summary by Maria Julissa Ek-Ramos) The Plant Cell. 10.1105/tpc.17.00537
Plants are exposed to a wide variety of biotic stressors such as herbivores and microbial pathogens. Thus, they have developed a sophisticated response system which involves different signal molecules and accurate signaling pathway responses. Among them, phytosulfokine (PSK), a disulfated pentapeptide, has been suggested to be a key signaling molecule, but evidence that supports this fact has been scarce. In this report, Zhang et al. show that PSK is a damage-associated signaling molecule with potential to be used to enhance disease resistance in different plant species. They show that PSK biosynthesis, modification and perception are very important for plant immunity related to tomato responses against the fungal pathogen Botrytis cinerea, and that PSK activation of the PSK receptor 1, RPSK1, enhances calcium cytosolic concentration. This increase in calcium content is sensed by the calcium binding protein calmodulin which then interacts with YUCs (auxin biosynthesis enzymes) initiating auxin-dependent immune responses. (Summary by Maria Julissa Ek-Ramos) The Plant Cell. 10.1105/tpc.17.00537
Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants
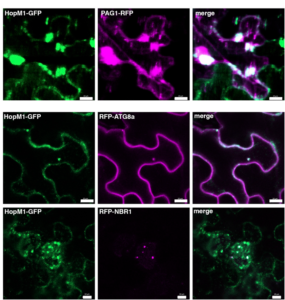 Autophagy has been defined as non-specific self-eating to obtain material that will be used for key processes within the cell. Even through authophagy has been demonstrated to be very important, its role during plant-bacteria interactions is not well known. Üstün et al. examined interactions between autophagy and the ubiquitin-proteasome system (UPS), based on previous studies showing the plant proteasome system is key to enhanced resistance against bacteria. Compromised proteasome systems are “recycled” by autophagy, therefore the authors suggested that if this process is compromised by bacterial infection, virulence is enhanced. They used pathogenic bacteria Pseudomonas syringae pv tomato DC 3000 and Arabidopsis thaliana as model systems. They found that bacterial infection enhances autophagy through bacterial effectors such as HopM1 in addition to the removal of proteasomes. However, as different effectors showed different results, they concluded that there should be different autophagy pathways that operate facilitating or limiting bacterial infections. In addition, one perspective of this work is to find more proteins that are targeted by the autophagy system in order to understand their role during pathogen infections. (Summary by Maria Julissa Ek-Ramos) The Plant Cell. 10.1105/tpc.17.00815
Autophagy has been defined as non-specific self-eating to obtain material that will be used for key processes within the cell. Even through authophagy has been demonstrated to be very important, its role during plant-bacteria interactions is not well known. Üstün et al. examined interactions between autophagy and the ubiquitin-proteasome system (UPS), based on previous studies showing the plant proteasome system is key to enhanced resistance against bacteria. Compromised proteasome systems are “recycled” by autophagy, therefore the authors suggested that if this process is compromised by bacterial infection, virulence is enhanced. They used pathogenic bacteria Pseudomonas syringae pv tomato DC 3000 and Arabidopsis thaliana as model systems. They found that bacterial infection enhances autophagy through bacterial effectors such as HopM1 in addition to the removal of proteasomes. However, as different effectors showed different results, they concluded that there should be different autophagy pathways that operate facilitating or limiting bacterial infections. In addition, one perspective of this work is to find more proteins that are targeted by the autophagy system in order to understand their role during pathogen infections. (Summary by Maria Julissa Ek-Ramos) The Plant Cell. 10.1105/tpc.17.00815


