What We’re Reading: March 9th
 Review. Plant evolution: landmarks on the path to terrestrial life
Review. Plant evolution: landmarks on the path to terrestrial life
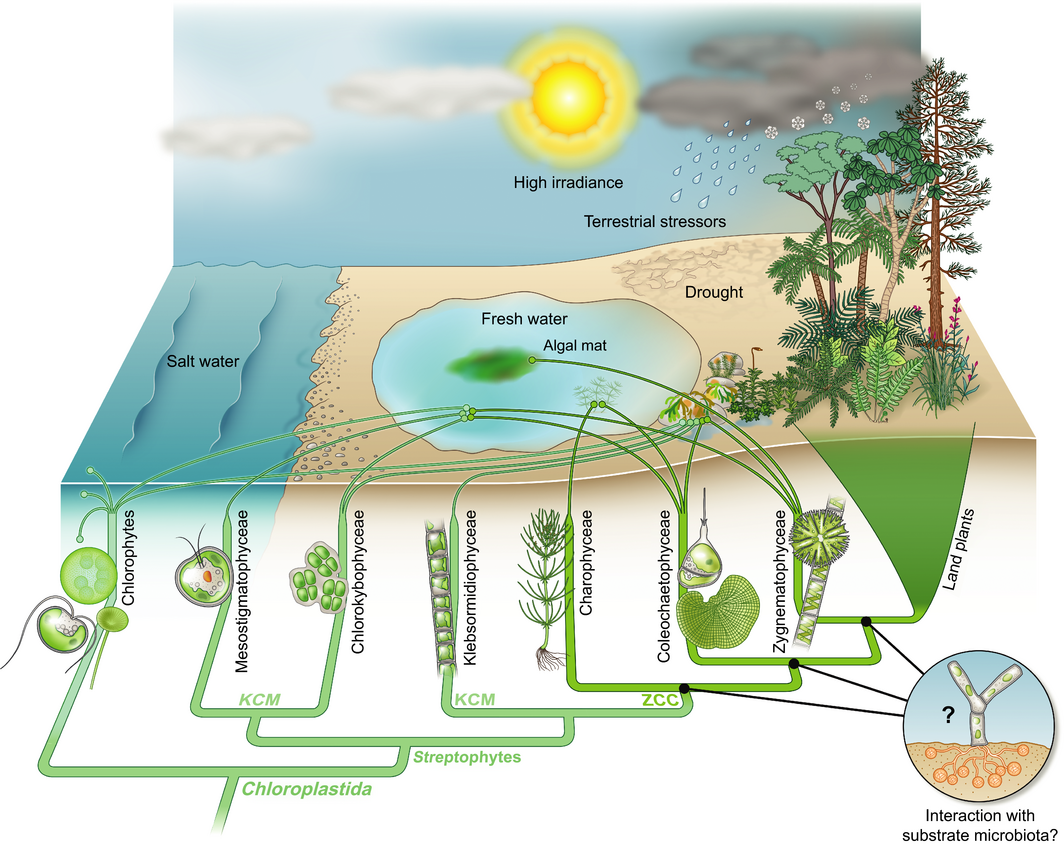
“Simply put, land plants evolved once; the biological significance of this singularity is writ large across the surface of the globe.” When I consider the incredible diversity found in life’s rich tapestry, I’m continually amazed that of all the different algae, only one lineage.is ancestral to all of the land plants; it just seems so improbable. How and why this happened is the focus of a new Tansley Insight by de Vries and Archibald, who review recent progress to understand this truly unique evolutionary event. The key features that enabled the algal embryophyte progenitor to colonize terrestrial habitats are described. The authors include a glossary of terms that define key land plant features as well as a beautiful illustration of the streptophyte lineage. (Summary by Mary Williams) New Phytol. 10.1111/nph.14975
High contiguity Arabidopsis thaliana genome assembly with a single nanopore flow cell
 Every now and then a technology arrives on the scene that suddenly makes everything easier. When I was a student, PCR was developed. More recently, CRISPR/Cas9 applications were developed, and now the hot tool is nanopore flow cell sequencing. The crucial advancement in this method is that rather than sequencing thousands of short sequences and having to find overlaps and paste them together, much longer strands (>200 kb) can be read. Michael et al. describe the use of this technology to sequence an Arabidopsis genome. They demonstrate that this method can “rapidly and inexpensively resolve structural variation (SV) at a quantitative trait locus (QTL) in A. thaliana.” (Summary by Mary Williams) Nature Comms. 10.1038/s41467-018-03016-2
Every now and then a technology arrives on the scene that suddenly makes everything easier. When I was a student, PCR was developed. More recently, CRISPR/Cas9 applications were developed, and now the hot tool is nanopore flow cell sequencing. The crucial advancement in this method is that rather than sequencing thousands of short sequences and having to find overlaps and paste them together, much longer strands (>200 kb) can be read. Michael et al. describe the use of this technology to sequence an Arabidopsis genome. They demonstrate that this method can “rapidly and inexpensively resolve structural variation (SV) at a quantitative trait locus (QTL) in A. thaliana.” (Summary by Mary Williams) Nature Comms. 10.1038/s41467-018-03016-2
High yield and early maturation in rice overexpressing nitrate transporter
 The application of nitrogen-containing fertilizers is crucial for good yields of crops like rice but also is energy intensive and polluting, so many approaches are being explored to develop plants that are more efficient in their uptake and use of nitrogen. Previous studies identified a crucial role of AtNRT1.1 (AtNPF6.3) as a nitrate sensor and transporter in Arabidopsis. Rice has three putative homologues of AtNRT1.1: OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C (OsNPF6.4). Wang et al. overexpressed one of these, OsNRT1.1A (from its own and a constitutively-expressed actin promoter), and found that the resulting plants showed improved utilization of N. Similar results were seen when this gene was overexpressed in Arabidopsis, showing the potential for it to enhance N utilization in both monocots and dicots. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00809
The application of nitrogen-containing fertilizers is crucial for good yields of crops like rice but also is energy intensive and polluting, so many approaches are being explored to develop plants that are more efficient in their uptake and use of nitrogen. Previous studies identified a crucial role of AtNRT1.1 (AtNPF6.3) as a nitrate sensor and transporter in Arabidopsis. Rice has three putative homologues of AtNRT1.1: OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C (OsNPF6.4). Wang et al. overexpressed one of these, OsNRT1.1A (from its own and a constitutively-expressed actin promoter), and found that the resulting plants showed improved utilization of N. Similar results were seen when this gene was overexpressed in Arabidopsis, showing the potential for it to enhance N utilization in both monocots and dicots. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00809
Transcriptomics of deepwater rice
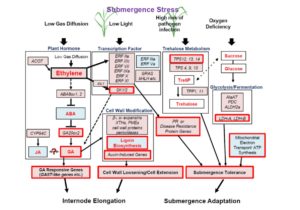 For most plants, becoming submerged under water can be lethal due to a restriction in gas exchange. One strategy for submergence tolerance is called an escape strategy, such as that employed by deepwater rice; the plant elongates rapidly to raise its leaves above the water level. Minami et al. used transcriptomics to further characterize the deepwater rice response. Key genes involved in the elongation response include those involved in “gibberellin biosynthesis, trehalose biosynthesis, anaerobic fermentation, cell wall modification and transcription factors that include ethylene-responsive factors.” The study also identified a role for jasmonic acid in internode elongation. Collectively, these transcriptional responses can protect against submergence stress by contributing to internode elongation, alternative energy production systems, and defense against pathogens. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.00858
For most plants, becoming submerged under water can be lethal due to a restriction in gas exchange. One strategy for submergence tolerance is called an escape strategy, such as that employed by deepwater rice; the plant elongates rapidly to raise its leaves above the water level. Minami et al. used transcriptomics to further characterize the deepwater rice response. Key genes involved in the elongation response include those involved in “gibberellin biosynthesis, trehalose biosynthesis, anaerobic fermentation, cell wall modification and transcription factors that include ethylene-responsive factors.” The study also identified a role for jasmonic acid in internode elongation. Collectively, these transcriptional responses can protect against submergence stress by contributing to internode elongation, alternative energy production systems, and defense against pathogens. (Summary by Mary Williams) Plant Physiol. 10.1104/pp.17.00858
Complete enzyme set for chlorophyll biosynthesis in Escherichia coli
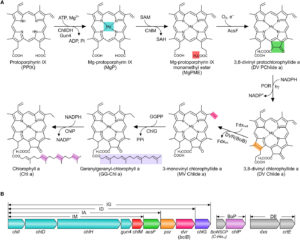 Although the reactions and enzymes involved in the biosynthesis of chlorophyll are well known, the entire pathway has never before been reconstituted in a non-photosynthetic organism. Chen et al. have done this. The cells (E. coli) expressing the full pathway accumulate chlorophyll and look green! However, they are not photosynthetic, as chlorophyll is just one of many components needed for photosynthesis. The authors report, “Our results delineate a minimum set of enzymes required to make chlorophyll and establish a platform for engineering photosynthesis in a heterotrophic model organism.” (Summary by Mary Williams) Sci. Adv. 10.1126/sciadv.aaq1407
Although the reactions and enzymes involved in the biosynthesis of chlorophyll are well known, the entire pathway has never before been reconstituted in a non-photosynthetic organism. Chen et al. have done this. The cells (E. coli) expressing the full pathway accumulate chlorophyll and look green! However, they are not photosynthetic, as chlorophyll is just one of many components needed for photosynthesis. The authors report, “Our results delineate a minimum set of enzymes required to make chlorophyll and establish a platform for engineering photosynthesis in a heterotrophic model organism.” (Summary by Mary Williams) Sci. Adv. 10.1126/sciadv.aaq1407
Crystal structure and pH dependency of peptidase & ligase activity in Arabidopsis legumain
 Legumains are proteins identified first from legumes that have subsequently been identified in other organisms. They are also described as asparaginyl endopeptidases (AEPs) and vacuolar cysteine proteases or vacuolar processing enzymes. Enzymatically, legumains can express peptidase and peptide ligase activities, and have been demonstrated to contribute to the formation of cyclic peptides. Because of the myriad potential applications for cyclic peptides as bioactive agents and pharmaceutical platforms, there is considerable interest in developing engineered, tunable enzymes for their production. Zauner et al. have identified the crystal structure of an Arabidopsis vegetative-type legumain, AtLEGγ, and show that its activity and specificity is strongly pH sensitive, supporting its putative functions in various compartments of the plant cell, but also providing an opportunity to control its activity in vitro. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00963
Legumains are proteins identified first from legumes that have subsequently been identified in other organisms. They are also described as asparaginyl endopeptidases (AEPs) and vacuolar cysteine proteases or vacuolar processing enzymes. Enzymatically, legumains can express peptidase and peptide ligase activities, and have been demonstrated to contribute to the formation of cyclic peptides. Because of the myriad potential applications for cyclic peptides as bioactive agents and pharmaceutical platforms, there is considerable interest in developing engineered, tunable enzymes for their production. Zauner et al. have identified the crystal structure of an Arabidopsis vegetative-type legumain, AtLEGγ, and show that its activity and specificity is strongly pH sensitive, supporting its putative functions in various compartments of the plant cell, but also providing an opportunity to control its activity in vitro. (Summary by Mary Williams) Plant Cell 10.1105/tpc.17.00963
A family of small, cyclic peptides buried in preproalbumin since the Eocene epoch
 Small cyclic peptides are produced in many organisms; some are produced by ribosomes and others not. Fisher et al. explore a class of small cyclic peptides known as orbitides, which have been identified in plants of Asteroideae subfamily of the plant family Asteraceae. The fact that these are abundant in but restricted to a single subfamily suggests that they evolved approximately 45 million years ago. The 5- to 12-amino acid long orbitides are ribosomally synthesized as part of a larger protein that also includes the seed-storage protein albumin. The authors previously used a transcriptomic approach to identify orbitide-encoding transcripts. In this work they sequenced the orbitides using liquid chromatography–coupled tandem mass spectroscopy (LC-MS/MS), and used NMR to determine the structures of several of them. In spite of the fact that many small cyclic peptides show antimicrobial activity, those in this study did not. (Summary by Mary Williams) Plant Direct 10.1002/pld3.42
Small cyclic peptides are produced in many organisms; some are produced by ribosomes and others not. Fisher et al. explore a class of small cyclic peptides known as orbitides, which have been identified in plants of Asteroideae subfamily of the plant family Asteraceae. The fact that these are abundant in but restricted to a single subfamily suggests that they evolved approximately 45 million years ago. The 5- to 12-amino acid long orbitides are ribosomally synthesized as part of a larger protein that also includes the seed-storage protein albumin. The authors previously used a transcriptomic approach to identify orbitide-encoding transcripts. In this work they sequenced the orbitides using liquid chromatography–coupled tandem mass spectroscopy (LC-MS/MS), and used NMR to determine the structures of several of them. In spite of the fact that many small cyclic peptides show antimicrobial activity, those in this study did not. (Summary by Mary Williams) Plant Direct 10.1002/pld3.42
Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis
 In plant cells, vacuoles store and degrade wastes including proteins. Vacuolar targeting and fusion by endosomes are specified by SNARE complexes at the membrane. In this study, Takemoto et al. studied the function of two tethering complexes, HOPS (Homotypic fusion and Protein Sorting) and CORVET (Class C core vacuole/endosome tethering), that had previously not been studied in plants. The authors showed that HOPS and CORVET mediate distinct membrane fusion events at the vacuole, and that their roles in plants are distinct from their roles in yeast and animal cells. HOPS acts with the VAMP713-containing SNARE complex and RAB7 to mediate membrane fusion between vacuoles, while CORVET mediates membrane fusion between multivesicular endosomes and the vacuole, in coordination with RAB5 and the VAMP727-containing SNARE complex. This work “provides further evidence for the unique evolutionary diversification of the vacuolar transport system in plants.” (Summary by Arif Ashraf) PNAS: 10.1073/pnas.1717839115
In plant cells, vacuoles store and degrade wastes including proteins. Vacuolar targeting and fusion by endosomes are specified by SNARE complexes at the membrane. In this study, Takemoto et al. studied the function of two tethering complexes, HOPS (Homotypic fusion and Protein Sorting) and CORVET (Class C core vacuole/endosome tethering), that had previously not been studied in plants. The authors showed that HOPS and CORVET mediate distinct membrane fusion events at the vacuole, and that their roles in plants are distinct from their roles in yeast and animal cells. HOPS acts with the VAMP713-containing SNARE complex and RAB7 to mediate membrane fusion between vacuoles, while CORVET mediates membrane fusion between multivesicular endosomes and the vacuole, in coordination with RAB5 and the VAMP727-containing SNARE complex. This work “provides further evidence for the unique evolutionary diversification of the vacuolar transport system in plants.” (Summary by Arif Ashraf) PNAS: 10.1073/pnas.1717839115
Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane
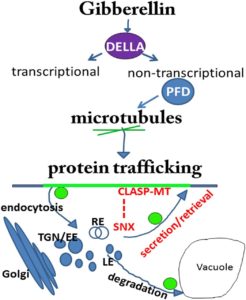 Gibberellins including gibberellic acid (GA) are plant hormones required for growth and development. GA has been reported to regulate tropic responses by modulating plasma membrane localization of PIN auxin transporters. In this study, Salanenka et al. focused on the mechanism of GA-mediated protein trafficking to the plasma membrane. They showed that low levels of GA promote delivery of PIN proteins to the vacuole for degradation. In contrast, high levels of GA promote the recycling of PIN proteins to the plasma membrane. This GA-mediated effect requires SNX1 (Sorting Nexin 1) and CLASP1 (Cytoplasmic Linker-Associated Protein 1). This study provides evidence that DELLA proteins function non-transcriptionally by regulating microtubule and retromer complexes through their interacting partners Prefoldins (PFDs), and that GA has the capacity to redirect auxin transporters to the plasma membrane and regulate PIN-dependent auxin fluxes. (Summary by Arif Ashraf) PNAS: 10.1073/pnas.1721760115
Gibberellins including gibberellic acid (GA) are plant hormones required for growth and development. GA has been reported to regulate tropic responses by modulating plasma membrane localization of PIN auxin transporters. In this study, Salanenka et al. focused on the mechanism of GA-mediated protein trafficking to the plasma membrane. They showed that low levels of GA promote delivery of PIN proteins to the vacuole for degradation. In contrast, high levels of GA promote the recycling of PIN proteins to the plasma membrane. This GA-mediated effect requires SNX1 (Sorting Nexin 1) and CLASP1 (Cytoplasmic Linker-Associated Protein 1). This study provides evidence that DELLA proteins function non-transcriptionally by regulating microtubule and retromer complexes through their interacting partners Prefoldins (PFDs), and that GA has the capacity to redirect auxin transporters to the plasma membrane and regulate PIN-dependent auxin fluxes. (Summary by Arif Ashraf) PNAS: 10.1073/pnas.1721760115
Why and how plants make puzzle cells
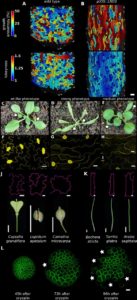 In proliferating tissues, plant cells start small and then expand to up to 100 times their original size. Sapala, Runions and collaborators studied the relationship between mechanical stress and shape to see if mechanical stress could affect the shape of epidermal cells. Simulations were run to see the effect of turgor-induced mechanical stress in single cells. When lobes were added to the cell, it was observed that the stress to the cell was not large and it was the best way to increase size without adding much stress to the cell. A mechanical model of puzzle shapes was created: cells will grow, stresses will increase and after a threshold the cell wall will be strengthened to resist these stresses. Puzzle shaped cells will appear when tissue grows isotropically (i.e equally in all directions). The model was tested with data from time lapse confocal imaging on cotyledons, showing a good correlation. Further analysis of data from other organs and species support the key role of mechanical stress in shape morphogenesis. (Summary by Cecilia Vasquez-Robinet) eLife 10.7554/eLife.32794.001
In proliferating tissues, plant cells start small and then expand to up to 100 times their original size. Sapala, Runions and collaborators studied the relationship between mechanical stress and shape to see if mechanical stress could affect the shape of epidermal cells. Simulations were run to see the effect of turgor-induced mechanical stress in single cells. When lobes were added to the cell, it was observed that the stress to the cell was not large and it was the best way to increase size without adding much stress to the cell. A mechanical model of puzzle shapes was created: cells will grow, stresses will increase and after a threshold the cell wall will be strengthened to resist these stresses. Puzzle shaped cells will appear when tissue grows isotropically (i.e equally in all directions). The model was tested with data from time lapse confocal imaging on cotyledons, showing a good correlation. Further analysis of data from other organs and species support the key role of mechanical stress in shape morphogenesis. (Summary by Cecilia Vasquez-Robinet) eLife 10.7554/eLife.32794.001
Regulation of postembryonic root organogenesis from Arabidopsis leaf tissue
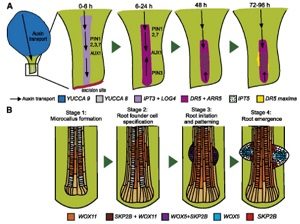 Organogenesis is an essential process in embryonic development. Additionally, plants can generate new organs, such as roots, from postembryonic tissues utilizing postembryonic pathways. Bustillo-Avendaño and colleagues investigated de novo root formation from Arabidopsis thaliana leaf tissue. They used microscopy, hormone treatments and mutant analysis to identify four main developmental stages and important regulatory factors. Initial cytokinin and auxin biosynthesis in the leaf is followed by long-distance auxin transport which leads to proliferation of xylem-associated tissues. Thus, cytokinin and auxin signaling appears to be critical for initial callus formation while stem cell regulators control organ initiation. This study provides a plant organ regeneration model through the identification of key stages and important regulators of de novo organogenesis from Arabidopsis tissue. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.17.00980
Organogenesis is an essential process in embryonic development. Additionally, plants can generate new organs, such as roots, from postembryonic tissues utilizing postembryonic pathways. Bustillo-Avendaño and colleagues investigated de novo root formation from Arabidopsis thaliana leaf tissue. They used microscopy, hormone treatments and mutant analysis to identify four main developmental stages and important regulatory factors. Initial cytokinin and auxin biosynthesis in the leaf is followed by long-distance auxin transport which leads to proliferation of xylem-associated tissues. Thus, cytokinin and auxin signaling appears to be critical for initial callus formation while stem cell regulators control organ initiation. This study provides a plant organ regeneration model through the identification of key stages and important regulators of de novo organogenesis from Arabidopsis tissue. (Summary by Julia Miller) Plant Physiol. 10.1104/pp.17.00980
Systemin receptor SYR1 enhances resistance against herbivorous insects
 Systemin is the first polypeptide hormone identified in plants, in 1991. Systemin is a small polypeptide hormone that conveys information about insect herbivory systemically (to other tissues). A previous report suggested that the tomato homologue of the brassinosteroid receptor BRI1 serves as a systemin receptor, but this finding has been hard to replicate. Now, starting with introgression lines between cultivated and wild tomato (systemin-responsive and non-responsive, respectively), Wang et al. mapped and identified SYR1 as a high-affinity, high-specificity systemin receptor. When SYR1 was introduced into Arabidopsis or tobacco, it conferred upon them sensitivity to systemin. When introduced into the non-responsive tomato species, it conferred resistance against insect herbivory. These studies demonstrate a role for SYR1 in systemin perception and defense against herbivory. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0106-0
Systemin is the first polypeptide hormone identified in plants, in 1991. Systemin is a small polypeptide hormone that conveys information about insect herbivory systemically (to other tissues). A previous report suggested that the tomato homologue of the brassinosteroid receptor BRI1 serves as a systemin receptor, but this finding has been hard to replicate. Now, starting with introgression lines between cultivated and wild tomato (systemin-responsive and non-responsive, respectively), Wang et al. mapped and identified SYR1 as a high-affinity, high-specificity systemin receptor. When SYR1 was introduced into Arabidopsis or tobacco, it conferred upon them sensitivity to systemin. When introduced into the non-responsive tomato species, it conferred resistance against insect herbivory. These studies demonstrate a role for SYR1 in systemin perception and defense against herbivory. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0106-0
The subgenomes of polyploid plants evolve at different rates
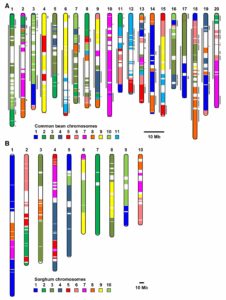 Polyploidy can result either through genome doubling followed by intraspecific crosses, or when two genomes from independent species hybridize, leading to autopolyploidy, or allopolyploidy, respectively. Polyploidy is an important evolutionary tool; the resultant redundancy from the duplication of genes allows for neofunctionalisation or subfunctionalisation to occur, which can lead to reproductive isolation, facilitating speciation and evolution. The evolutionary fate of subgenomes is dependent on whether the polyploid is the result of autopolyploidy or allopolyploidy, and how different the initial genomes where from one another. A recent study by Zhao et al. investigated the factors influencing the evolution of duplicated gene pairs in polyploids by comparing genomic and epigenomic differences in the genomes of maize and soybean. Maize likely resulted from an allopolyploidy event, and because of the initial differences between the two progenitor genomes, one subgenome exhibits dominance in gene expression and selective constraints over the other, influencing how the genome changes over time. Soybean, conversely, shows little evidence for subgenome dominance, and was, therefore, likely the result of an autopolyploidy, or an allopolyploidy from two closely related species, resulting in the soybean genome retaining the majority of duplicate genes. This study sheds light on the factors driving genome evolution, and what effects subgenome dominance can have over long time periods. (Summary by Danielle Roodt Prinsloo) Plant Cell 10.1105/tpc.17.00595
Polyploidy can result either through genome doubling followed by intraspecific crosses, or when two genomes from independent species hybridize, leading to autopolyploidy, or allopolyploidy, respectively. Polyploidy is an important evolutionary tool; the resultant redundancy from the duplication of genes allows for neofunctionalisation or subfunctionalisation to occur, which can lead to reproductive isolation, facilitating speciation and evolution. The evolutionary fate of subgenomes is dependent on whether the polyploid is the result of autopolyploidy or allopolyploidy, and how different the initial genomes where from one another. A recent study by Zhao et al. investigated the factors influencing the evolution of duplicated gene pairs in polyploids by comparing genomic and epigenomic differences in the genomes of maize and soybean. Maize likely resulted from an allopolyploidy event, and because of the initial differences between the two progenitor genomes, one subgenome exhibits dominance in gene expression and selective constraints over the other, influencing how the genome changes over time. Soybean, conversely, shows little evidence for subgenome dominance, and was, therefore, likely the result of an autopolyploidy, or an allopolyploidy from two closely related species, resulting in the soybean genome retaining the majority of duplicate genes. This study sheds light on the factors driving genome evolution, and what effects subgenome dominance can have over long time periods. (Summary by Danielle Roodt Prinsloo) Plant Cell 10.1105/tpc.17.00595


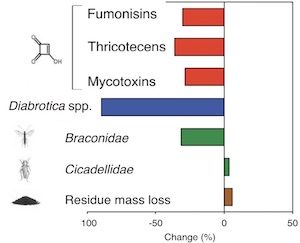 The question of the relative risks and benefits of genetically engineered (GE) maize is still in the middle of a heated debate, despite the widespread cultivation of GE crops (12% of the global crop-land) and long standing commercialization (since 1996). Pellegrino et al. report a meta-analysis of the existing data on the GE maize related agronomic, environmental and toxicological impacts reported in the scientific literature between the years 1996 and 2016. The authors conclude that their analysis illustrates that GE maize shows benefits in terms of increases in grain yield and quality, and in decreases of the trans-gene targetted insects when compared to conventional corn varieties. Their analysis points towards a small or possibly no effect of GE maize on the abundance of non-targeted insects, and provides evidence that GE corn cultivation reduces levels of harmful mycotoxin (−28.8%) content in resulting grain. (Summary by
The question of the relative risks and benefits of genetically engineered (GE) maize is still in the middle of a heated debate, despite the widespread cultivation of GE crops (12% of the global crop-land) and long standing commercialization (since 1996). Pellegrino et al. report a meta-analysis of the existing data on the GE maize related agronomic, environmental and toxicological impacts reported in the scientific literature between the years 1996 and 2016. The authors conclude that their analysis illustrates that GE maize shows benefits in terms of increases in grain yield and quality, and in decreases of the trans-gene targetted insects when compared to conventional corn varieties. Their analysis points towards a small or possibly no effect of GE maize on the abundance of non-targeted insects, and provides evidence that GE corn cultivation reduces levels of harmful mycotoxin (−28.8%) content in resulting grain. (Summary by