What We’re Reading: June 2nd
For the remainder of June and July, several Plantae Fellows will take turns editing the What We’re Reading series. The Plantae Fellows were selected for their interest in communicating science, and they are looking forward to wearing the Editor hat and sharing their selections with you in the coming weeks. (Note – if you are interested in applying to be a 2017-2018 Plantae Fellow, we will be issuing an application call in the next few days. Watch this space!)
Weekly “What We’re Reading” Guest Editors
June 9 Sridhar Gutam, Senior Scientist, Indian Council of Agricultural Research and Nidhi Sharma, Research Specialist, Stanford University
June 16 Sudhakar Srivastava, Postdoctoral Fellow at Blaustein Institute for Desert Research, Ben-Gurion University, Israel
June 23 Gaby Auge, Senior Researcher at Fundación Insituto Leloir, IIBBA-CONICET, Argentina
July 30 Mather Khan, Postdoctoral Fellow, University of Missouri
July 7 KK Sabu, Senior Scientist, Jawaharlal Nehru Tropical Botanic Garden and Research Institute, India
July 21 Khalid Hakeem, Associate Professor, King Abdulazis University, Saudi Arabia
July 28 Bhavisha Sheth, Postdoctoral Fellow, Policy Research in Science, Technology and Innovation, India
Review: Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria ($)
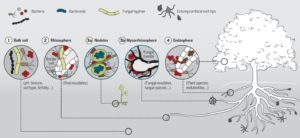 Martin et al. have written a must-read review on the evolution of plant mutualistic symbioses and strategies through which hosts and microbes communicate and coordinate their activities. This review also covers the contributions of hormones in the formation of symbiotic tissues, and how the mutualistic microbes evade host defenses. The authors observe the need to scale up our understanding of host-mutualist relations to encompass numerous interactions occurring simultaneously, and to integrate how nutrient trade dynamics are regulated. They also discuss how understanding these interactions can lead to enhanced agricultural productivity. Science 10.1126/science.aad4501 Tags: Biotic Interactions, Evolution
Martin et al. have written a must-read review on the evolution of plant mutualistic symbioses and strategies through which hosts and microbes communicate and coordinate their activities. This review also covers the contributions of hormones in the formation of symbiotic tissues, and how the mutualistic microbes evade host defenses. The authors observe the need to scale up our understanding of host-mutualist relations to encompass numerous interactions occurring simultaneously, and to integrate how nutrient trade dynamics are regulated. They also discuss how understanding these interactions can lead to enhanced agricultural productivity. Science 10.1126/science.aad4501 Tags: Biotic Interactions, Evolution
Review: Auxin perception and downstream events ($)
 The mechanism by which auxin alters patters of transcription through TIR1, ARFs and IAA proteins is well established, but less clear is our understanding of how rapid, non-transcriptional auxin responses occur. APB1 was implicated in non-transcriptional responses, but in the past few years evidence for ABP1 involvement has been challenged. Strader and Zhao review the current status of ABP1 research. Students should be encouraged to read this review given the important lessons that may be learned from the rise and fall of ABP1. The authors also propose a model in which PIN proteins act as auxin receptors through a signaling pathway similar to that of the PHOT1 blue light response. Curr. Opin. Plant Biol. 10.1016/j.pbi.2016.04.004 Tags: Growth Regulation, Education and Outreach, Signals and Responses
The mechanism by which auxin alters patters of transcription through TIR1, ARFs and IAA proteins is well established, but less clear is our understanding of how rapid, non-transcriptional auxin responses occur. APB1 was implicated in non-transcriptional responses, but in the past few years evidence for ABP1 involvement has been challenged. Strader and Zhao review the current status of ABP1 research. Students should be encouraged to read this review given the important lessons that may be learned from the rise and fall of ABP1. The authors also propose a model in which PIN proteins act as auxin receptors through a signaling pathway similar to that of the PHOT1 blue light response. Curr. Opin. Plant Biol. 10.1016/j.pbi.2016.04.004 Tags: Growth Regulation, Education and Outreach, Signals and Responses
Review: Phosphate scouting by root tips ($)
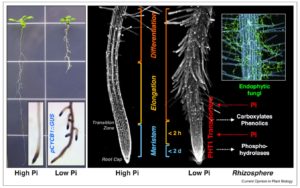 Phosphate is both really important (think of its abundance in DNA, RNA, ATP, and membrane lipids), and really difficult to assimilate due to its insolubility and immobility in soil. Therefore, phosphate is frequently limiting for growth and widely applied as fertilizer, but global supplies of phosphate are finite and the raw material becoming increasingly difficult to obtain (see this for example). As a consequence, there is considerable interest in identifying plant responses to limiting phosphate, including as reviewed by Abel the strategies by which roots sense and respond to phosphate. Phosphate assimilation is complicated by the presence of metallic cations Fe3+ and Al3+, and the interactions between low phosphate and the uptake of these ions is also discussed. Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.04.016 Tags: Ecophysiology, Physiology, Transport
Phosphate is both really important (think of its abundance in DNA, RNA, ATP, and membrane lipids), and really difficult to assimilate due to its insolubility and immobility in soil. Therefore, phosphate is frequently limiting for growth and widely applied as fertilizer, but global supplies of phosphate are finite and the raw material becoming increasingly difficult to obtain (see this for example). As a consequence, there is considerable interest in identifying plant responses to limiting phosphate, including as reviewed by Abel the strategies by which roots sense and respond to phosphate. Phosphate assimilation is complicated by the presence of metallic cations Fe3+ and Al3+, and the interactions between low phosphate and the uptake of these ions is also discussed. Curr. Opin. Plant Biol. 10.1016/j.pbi.2017.04.016 Tags: Ecophysiology, Physiology, Transport
Exploring the chemical diversity and distribution of marine cyanobacteria and algae through mass spectrometry
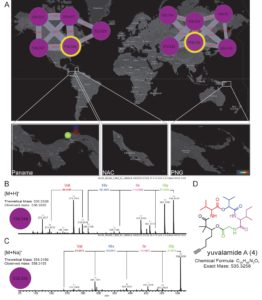 Like plants, algae and cyanobacteria produce a dizzying array of largely unexplored natural products that can be used as medicines, in biotechnology and in agriculture. Luzzatto-Knaan et al. used mass spectrometry (MS) to explore chemical diversity from marine samples collected across space and time. They found some compounds that are widely dispersed and others found only in specific regions, and they displayed their findings on an interactive world map, revealing regions of high chemical diversity. From a collection obtained from one of these regions, they identified a novel natural product, yuvalamide A. This work demonstrates the value of using non-targeted MS approaches to characterize metabolites, as well as highlights the uncharted information stored in existing collections. eLIFE 10.7554/eLife.24214 Tags: Biochemistry, Ecology, Metabolism
Like plants, algae and cyanobacteria produce a dizzying array of largely unexplored natural products that can be used as medicines, in biotechnology and in agriculture. Luzzatto-Knaan et al. used mass spectrometry (MS) to explore chemical diversity from marine samples collected across space and time. They found some compounds that are widely dispersed and others found only in specific regions, and they displayed their findings on an interactive world map, revealing regions of high chemical diversity. From a collection obtained from one of these regions, they identified a novel natural product, yuvalamide A. This work demonstrates the value of using non-targeted MS approaches to characterize metabolites, as well as highlights the uncharted information stored in existing collections. eLIFE 10.7554/eLife.24214 Tags: Biochemistry, Ecology, Metabolism
Identification of putative substrates of SEC2, a chloroplast inner envelope translocase
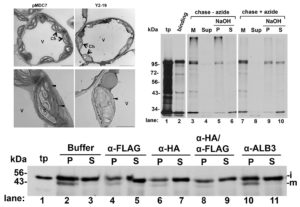
Chloroplasts have evolved from photosynthetic bacteria, and genes necessary for chloroplast function have moved from the chloroplast to the nuclear genome of the host eukaryotic cell. Proteins encoded by these genes are made in the cytosol and imported into the chloroplast using protein translocases. Chloroplasts contain the characterized SEC1 and the recently-discovered SEC2 translocase systems. The SEC2 translocase consists of the SECA2, SCY2, and SECE2 proteins. Since SEC2 was recently discovered, its protein substrates are unknown. Inducible RNAi targeting of SCY2 revealed decreased levels of many candidate proteins upon silencing. To identify substrates for SEC2, Li, et al. developed an in vitro method to test protein integration into isolated chloroplast membranes. Using this assay, FTSH12 integration in the assay was inhibited by the SECA inhibitor sodium azide, which identified FTSH12 as a SEC2 substrate. TIC40 was identified as a second substrate because antibodies against epitope-tagged SCY2 and SECE2 inhibited TIC40 integration. (Review by Daniel Czerny) Plant Physiol. Tags: Cell Biology
Dehydration stress extends mRNA 3′ UTRs with noncoding RNA functions ($)
 Sun et al. observed that under dehydration stress, many genes showed a 3′ untranslated region (3′ UTR) extension of roughly 200 – 800 nucleotides. The outcome of these extensions appears to be the regulation of other genes. For example, through their extension, many (more than 600) of the extended mRNAs overlapped with genes on the opposite strand (described here as sense transcripts), and many of these sense transcript genes showed a decrease in expression levels. Other extended RNAs overlapped with promoters of genes on the same strand via their extension, resulting in an increase in expression levels of the overlapped genes. This report suggests a novel strategy for stress-regulated gene expression. Genome Research 10.1101/gr.218669.116 Another paper published this week, by de Lorenzo et al., shows that alternate polyadenylation sites affect mRNA stability and so gene expression during hypoxia – see Plant Cell 10.1105/tpc.16.00746 Tags: Abiotic Interactions, Gene Regulation, Molecular Biology
Sun et al. observed that under dehydration stress, many genes showed a 3′ untranslated region (3′ UTR) extension of roughly 200 – 800 nucleotides. The outcome of these extensions appears to be the regulation of other genes. For example, through their extension, many (more than 600) of the extended mRNAs overlapped with genes on the opposite strand (described here as sense transcripts), and many of these sense transcript genes showed a decrease in expression levels. Other extended RNAs overlapped with promoters of genes on the same strand via their extension, resulting in an increase in expression levels of the overlapped genes. This report suggests a novel strategy for stress-regulated gene expression. Genome Research 10.1101/gr.218669.116 Another paper published this week, by de Lorenzo et al., shows that alternate polyadenylation sites affect mRNA stability and so gene expression during hypoxia – see Plant Cell 10.1105/tpc.16.00746 Tags: Abiotic Interactions, Gene Regulation, Molecular Biology
Role of SPA proteins in COP1 subcellular localization ($)
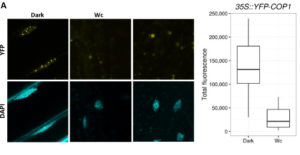 SPA (SUPPRESSOR OF PHYTOCHROME A-105) proteins form a complex with COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) and are required for COP1 to respond to changes in the light environment and transduce light signals and influence plant development. In this paper, Balcerowicz et al. investigated the importance of SPA proteins in the COP1/SPA complex. Using fluorescence, they could locate exactly where COP1 was in seedlings grown in light or kept in darkness, in a background with or without SPA proteins. Their results show that SPA proteins are not necessary for COP1 to localize in the nucleus but they are required for the complex to leave the nucleus after exposure to light, without affecting its degradation rate. Furthermore, they show that COP1 localization mediated by SPA proteins respond differently to different light wavelengths. Altogether, the results show that SPA proteins are necessary for COP1 responsiveness to changes in the light environment by altering its subcellular localization and, in consequence, the proper response of plants to light. (Review by Gaby Auge) Plant Physiology, 10.1104/pp.17.00488 Tags: Cell Biology, Signals and Responses
SPA (SUPPRESSOR OF PHYTOCHROME A-105) proteins form a complex with COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1) and are required for COP1 to respond to changes in the light environment and transduce light signals and influence plant development. In this paper, Balcerowicz et al. investigated the importance of SPA proteins in the COP1/SPA complex. Using fluorescence, they could locate exactly where COP1 was in seedlings grown in light or kept in darkness, in a background with or without SPA proteins. Their results show that SPA proteins are not necessary for COP1 to localize in the nucleus but they are required for the complex to leave the nucleus after exposure to light, without affecting its degradation rate. Furthermore, they show that COP1 localization mediated by SPA proteins respond differently to different light wavelengths. Altogether, the results show that SPA proteins are necessary for COP1 responsiveness to changes in the light environment by altering its subcellular localization and, in consequence, the proper response of plants to light. (Review by Gaby Auge) Plant Physiology, 10.1104/pp.17.00488 Tags: Cell Biology, Signals and Responses
Wild tobacco genomes reveal the evolution of nicotine biosynthesis
 The genus Nicotiana encompasses several species and hybrids, the most famous being Nicotiana tabacum, cultivated for production of tobacco. Xu et al. sequenced the genome of Nicotiana attenuata and Nicotiana obtusifolia with an interest in identifying the origins of nicotine biosynthesis. Nicotine is a toxic alkaloid produced to protect against herbivory, but also the addictive compound in tobacco products. Analysis of these genomes revealed that nicotine synthesis evolved in Nicotiana from duplication in the polyamine and nicotinamide adenine dinucleotide pathways, as well as proliferation of transposable elements. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1700073114 Tags: Biochemistry, Biotic Interactions, Evolution, Metabolism
The genus Nicotiana encompasses several species and hybrids, the most famous being Nicotiana tabacum, cultivated for production of tobacco. Xu et al. sequenced the genome of Nicotiana attenuata and Nicotiana obtusifolia with an interest in identifying the origins of nicotine biosynthesis. Nicotine is a toxic alkaloid produced to protect against herbivory, but also the addictive compound in tobacco products. Analysis of these genomes revealed that nicotine synthesis evolved in Nicotiana from duplication in the polyamine and nicotinamide adenine dinucleotide pathways, as well as proliferation of transposable elements. Proc. Natl. Acad. Sci. USA 10.1073/pnas.1700073114 Tags: Biochemistry, Biotic Interactions, Evolution, Metabolism
Bypassing negative epistasis on yield in tomato imposed by a domestication gene
 Tomatoes normally produce multiple flowers along a single branch. Soyk et al. explored a large collection of wild and domesticated accessions to identify those with branched inflorescences, which should be able to produce more fruit per plant. They identified a few related branchy mutants which they called s2, that are highly branching but with low fertility so fail to form many fruit. They subsequently recognized that the s2 phenotype arises from variation in two unlinked genes, J2 and EJ2, both of which encode MADS box transcription factors. The ej2w allele increases fruit size, and was probably selected for during domestication. The j2TE allele provides a desirable jointless phenotype that prevents fruit from dropping, but when combined with ej2w leads to excessive branching and decreased fruit yield. By separating out these two loci and engineering new alleles, tomatoes with more but not too much more branching and higher yelds were produced. Cell 10.1016/j.cell.2017.04.032 Tags: Applied Plant Biology, Development, Genetics
Tomatoes normally produce multiple flowers along a single branch. Soyk et al. explored a large collection of wild and domesticated accessions to identify those with branched inflorescences, which should be able to produce more fruit per plant. They identified a few related branchy mutants which they called s2, that are highly branching but with low fertility so fail to form many fruit. They subsequently recognized that the s2 phenotype arises from variation in two unlinked genes, J2 and EJ2, both of which encode MADS box transcription factors. The ej2w allele increases fruit size, and was probably selected for during domestication. The j2TE allele provides a desirable jointless phenotype that prevents fruit from dropping, but when combined with ej2w leads to excessive branching and decreased fruit yield. By separating out these two loci and engineering new alleles, tomatoes with more but not too much more branching and higher yelds were produced. Cell 10.1016/j.cell.2017.04.032 Tags: Applied Plant Biology, Development, Genetics
Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency
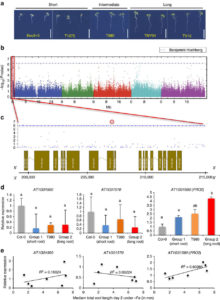 Iron is an essential nutrient that plants assimilate from the soil. Moderate iron deficiency induces an increase in primary root length and lateral root production. Satbhai et al. examined natural variation of root responses and showed a correlation between root length and allelic variation at the FRO2 locus which encodes a membrane-bound ferric chelate reductase involved in the reduction of Fe3+ to Fe2+ prior to uptake into the root. Specifically, elevated expression levels of FRO2 are correlated with longer roots in low iron conditions. Nature Comms. 10.1038/ncomms15603 Tags: Genetics, Physiology
Iron is an essential nutrient that plants assimilate from the soil. Moderate iron deficiency induces an increase in primary root length and lateral root production. Satbhai et al. examined natural variation of root responses and showed a correlation between root length and allelic variation at the FRO2 locus which encodes a membrane-bound ferric chelate reductase involved in the reduction of Fe3+ to Fe2+ prior to uptake into the root. Specifically, elevated expression levels of FRO2 are correlated with longer roots in low iron conditions. Nature Comms. 10.1038/ncomms15603 Tags: Genetics, Physiology
Monoterpenes support systemic acquired resistance within and between plants ($)
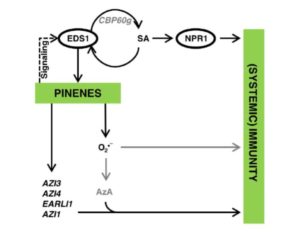 Pathogen perception leads to local and systemic immune responses including systemic acquired resistance (SAR). The nature of the mobile signals and their movements remain uncertain. Riedlmeier et al. demonstrated that certain monoterpenes including α- and β-pinene accumulate in SAR-inducing conditions and enhance systemic resistance. Futher, they show evidence for “a tight association between plant-to-plant SAR-like signaling and monoterpene emissions.” Plant Cell Tags: Biotic Interactions, Ecophysiology, Signals and Responses
Pathogen perception leads to local and systemic immune responses including systemic acquired resistance (SAR). The nature of the mobile signals and their movements remain uncertain. Riedlmeier et al. demonstrated that certain monoterpenes including α- and β-pinene accumulate in SAR-inducing conditions and enhance systemic resistance. Futher, they show evidence for “a tight association between plant-to-plant SAR-like signaling and monoterpene emissions.” Plant Cell Tags: Biotic Interactions, Ecophysiology, Signals and Responses
The pPSU plasmids for generating DNA molecular weight markers
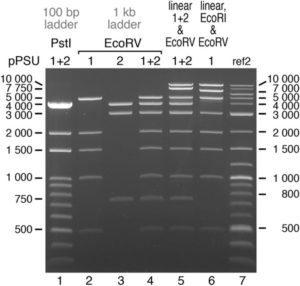 Anyone who is trying to manage a laboratory on a budget will enjoy reading about the efforts of Henrici et al., who have designed a pair of plasmids that together can be digested with one of two restriction enzymes to produce either 100-bp or 1000-bp DNA molecular weight markers. As the authors report, “We would like to offer the pPSU1 & pPSU2 pair of plasmids as an alternative to commercial or other homemade sources of DNA ladders. We therefore make available the pPSU1 and pPSU2 plasmids to the nonprofit academic community without licensing requirements. The pPSU1 and pPSU2 plasmids are being deposited in the Addgene and DNASU repositories to facilitate distribution.” Sci. Reports 10.1038/s41598-017-02693-1 Tags: Molecular Biology
Anyone who is trying to manage a laboratory on a budget will enjoy reading about the efforts of Henrici et al., who have designed a pair of plasmids that together can be digested with one of two restriction enzymes to produce either 100-bp or 1000-bp DNA molecular weight markers. As the authors report, “We would like to offer the pPSU1 & pPSU2 pair of plasmids as an alternative to commercial or other homemade sources of DNA ladders. We therefore make available the pPSU1 and pPSU2 plasmids to the nonprofit academic community without licensing requirements. The pPSU1 and pPSU2 plasmids are being deposited in the Addgene and DNASU repositories to facilitate distribution.” Sci. Reports 10.1038/s41598-017-02693-1 Tags: Molecular Biology
Rapid cytosolic calcium elevations in Arabidopsis during aphid feeding
 Calcium signaling is a common plant response to many different stimuli. Vincent et al. used a fluorescent calcium reporter, GCaMP3, to record calcium responses in Arabidopsis to aphid feeding (specifically, the green peach aphid Myzus persicae). Through analysis of various mutants, key components of the aphid calcium response were identified and a model proposed. The response is abolished in mutants of the co-receptor BAK1, and also in mutants of the plasma membrane cation-permeable channels GLR3.3 and GLR3.6. The calcium-permeable vacuolar channel TWO-PORE CHANNEL 1 (TPC1) also has a role; the signal is attenuated in loss-of-function mutants, and systemically enhanced (with a concommitant loss of aphid fecundity) in over-activation lines. These results demonstrate the importance of calcium signaling in plant responses to aphids, and suggest a potential strategy for insect resistance. Plant Cell 10.1105/tpc.17.00136 Tags: Biotic Interactions, Molecular Biology, Signals and Responses
Calcium signaling is a common plant response to many different stimuli. Vincent et al. used a fluorescent calcium reporter, GCaMP3, to record calcium responses in Arabidopsis to aphid feeding (specifically, the green peach aphid Myzus persicae). Through analysis of various mutants, key components of the aphid calcium response were identified and a model proposed. The response is abolished in mutants of the co-receptor BAK1, and also in mutants of the plasma membrane cation-permeable channels GLR3.3 and GLR3.6. The calcium-permeable vacuolar channel TWO-PORE CHANNEL 1 (TPC1) also has a role; the signal is attenuated in loss-of-function mutants, and systemically enhanced (with a concommitant loss of aphid fecundity) in over-activation lines. These results demonstrate the importance of calcium signaling in plant responses to aphids, and suggest a potential strategy for insect resistance. Plant Cell 10.1105/tpc.17.00136 Tags: Biotic Interactions, Molecular Biology, Signals and Responses



Leave a Reply
Want to join the discussion?Feel free to contribute!