What We’re Reading: August 3rd
Review: MYBs drive novel consumer traits in fruits and vegetables
 The MYB transcription factors, specifically the R2R3 family of MYBs, are closely associated with the regulation of anthocyanin biosynthesis. This easy-to-score trait made MYBs some of the earliest characterized plant transcription factors. Allan and Espley summarize the contributions of MYBs to pigmentation in flowers, fruits, leaves and even other organs (e.g., purple cauliflower). Because anthocyanins and related compounds have demonstrated health benefits, some of this knowledge is being used to increase their accumulation in food, for example in the flesh of fruits rather than only the skins. Furthermore, MYBs can also affect flavor, through for example production of tannins. Other roles for MYBs are also addressed; for example, the smooth-skinned nectarine differs from the fuzzy peach by a single MYB gene. This is an interesting and relatively accessible article that would be enjoyed by students. As the authors say, “MYBs are often the gene providing the ‘Wow Factor’. ” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.06.001
The MYB transcription factors, specifically the R2R3 family of MYBs, are closely associated with the regulation of anthocyanin biosynthesis. This easy-to-score trait made MYBs some of the earliest characterized plant transcription factors. Allan and Espley summarize the contributions of MYBs to pigmentation in flowers, fruits, leaves and even other organs (e.g., purple cauliflower). Because anthocyanins and related compounds have demonstrated health benefits, some of this knowledge is being used to increase their accumulation in food, for example in the flesh of fruits rather than only the skins. Furthermore, MYBs can also affect flavor, through for example production of tannins. Other roles for MYBs are also addressed; for example, the smooth-skinned nectarine differs from the fuzzy peach by a single MYB gene. This is an interesting and relatively accessible article that would be enjoyed by students. As the authors say, “MYBs are often the gene providing the ‘Wow Factor’. ” (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2018.06.001
Roles of N-terminal acetylation and N-terminal acetyltransferases in plants
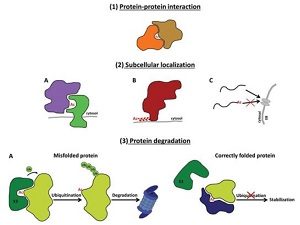 N-terminal acetylation is a common protein modification completed by ribosome-associated N-terminal acetyltransferases (NATs) or plastid-localized NATs. The recent discovery of the plastid-localized NATs upended the traditional model of static, co-translational imprinting of the plant proteome. These dynamic changes, currently not identified in other eukaryotes, could be due to the sessile habit of plants. NTAs have roles in protein-protein interactions, subcellular localization, folding and degradation, among other essential functions. This paper reviews plant NTAs, including their substrate specificity, their roles in plastids and the consequences of acetylation. The paper also discusses the potential roles of NTAs in abiotic and biotic stress responses. (Summary by Julia Miller) J. Exp. Bot. 10.1093/jxb/ery241
N-terminal acetylation is a common protein modification completed by ribosome-associated N-terminal acetyltransferases (NATs) or plastid-localized NATs. The recent discovery of the plastid-localized NATs upended the traditional model of static, co-translational imprinting of the plant proteome. These dynamic changes, currently not identified in other eukaryotes, could be due to the sessile habit of plants. NTAs have roles in protein-protein interactions, subcellular localization, folding and degradation, among other essential functions. This paper reviews plant NTAs, including their substrate specificity, their roles in plastids and the consequences of acetylation. The paper also discusses the potential roles of NTAs in abiotic and biotic stress responses. (Summary by Julia Miller) J. Exp. Bot. 10.1093/jxb/ery241
Review: Regulation of pattern recognition receptor signalling by phosphorylation and ubiquitination
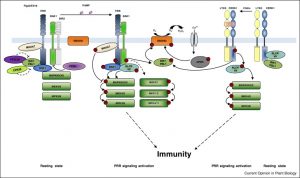 A key step in pathogen recognition occurs at the cell surface with the interaction between receptor-like kinases (pattern-recognition receptors) and their ligands. This event triggers a signal-transduction cascade that leads to the induction of defense responses. Mithoe and Menke review our current understanding of these signals, focusing on the contributions of protein activation and inactivation by phosphorylation and ubiquitination, largely uncovered through proteomics methods, and drawing on both rice and Arabidopsis models. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.07.008
A key step in pathogen recognition occurs at the cell surface with the interaction between receptor-like kinases (pattern-recognition receptors) and their ligands. This event triggers a signal-transduction cascade that leads to the induction of defense responses. Mithoe and Menke review our current understanding of these signals, focusing on the contributions of protein activation and inactivation by phosphorylation and ubiquitination, largely uncovered through proteomics methods, and drawing on both rice and Arabidopsis models. (Summary by Mary Williams) Curr. Opin. Plant Biol. 10.1016/j.pbi.2018.07.008
Opinion: What is the next frontier of plant engineering? ($)
 Cell asked six plants scientist to speculate on the next big application in plant science. The six scientists, Jeff Dangle, Anne Osbourn, Diego Orzáez, Zhikang Li, Stephen Long, and Julian Schroeder, each draw on their own expertise and interest in their answers, which include: applications towards disease resistance, designer metabolites, synthetic signalling circuits, breeding by design, improved photosynthesis, and greater water use efficiency. (Summary by Mary Williams) Cell 10.1016/j.cell.2018.07.012 (Image credit iAGRI Tanzania)
Cell asked six plants scientist to speculate on the next big application in plant science. The six scientists, Jeff Dangle, Anne Osbourn, Diego Orzáez, Zhikang Li, Stephen Long, and Julian Schroeder, each draw on their own expertise and interest in their answers, which include: applications towards disease resistance, designer metabolites, synthetic signalling circuits, breeding by design, improved photosynthesis, and greater water use efficiency. (Summary by Mary Williams) Cell 10.1016/j.cell.2018.07.012 (Image credit iAGRI Tanzania)
Diffusion of CO2 across the mesophyll-bundle sheath cell interface in a C4 plant with genetically reduced PEP carboxylase activity
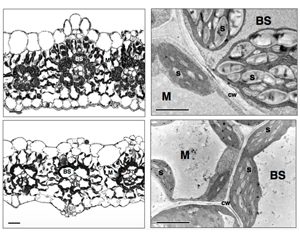 C4 photosynthesis relies on the transport of carbon (in the form of C4 acids) from the mesophyll into bundle sheath cells (BSCs). Subsequent decarboxylation of these C4 acids generates a high concentration of CO2 in the vicinity of Rubisco, helping to improve the catalytic efficiency of this enzyme. However, carbon can also enter BSCs via direct CO2 diffusion from the mesophyll – the passage of CO2 across this interface is termed bundle sheath conductance (gbs). In this research, Alonso-Cantabrana et al. used RNAi to target mesophyll phosphoenolpyruvate carboxylase (PEPC) in Setaria viridis. PEPC performs the initial fixation of CO2 from the air in the C4 system, and so in its absence, direct diffusion represents the major source of CO2 for Rubisco in BSCs. In the resulting plants, the authors were able to determine gbs, and also examine the mesophyll-BSC interface to establish the anatomical response to the introduced lesion. Gas exchange data showed that in the absence of PEPC, Rubisco appeared to be fixing CO2 directly, while plasmodesmatal density was increased in the low-PEPC plants, potentially as a compensation mechanism to allow higher CO2 flux into BSCs. This also represents a route for increased CO2 leakage from BSCs, which would normally be recycled by PEPC in the mesophyll. Investigation of the signalling pathways involved in this response represents a challenging next step in determining how gbs is modulated. (Summary by Mike Page) Plant Physiol. 10.1104/pp.18.00618
C4 photosynthesis relies on the transport of carbon (in the form of C4 acids) from the mesophyll into bundle sheath cells (BSCs). Subsequent decarboxylation of these C4 acids generates a high concentration of CO2 in the vicinity of Rubisco, helping to improve the catalytic efficiency of this enzyme. However, carbon can also enter BSCs via direct CO2 diffusion from the mesophyll – the passage of CO2 across this interface is termed bundle sheath conductance (gbs). In this research, Alonso-Cantabrana et al. used RNAi to target mesophyll phosphoenolpyruvate carboxylase (PEPC) in Setaria viridis. PEPC performs the initial fixation of CO2 from the air in the C4 system, and so in its absence, direct diffusion represents the major source of CO2 for Rubisco in BSCs. In the resulting plants, the authors were able to determine gbs, and also examine the mesophyll-BSC interface to establish the anatomical response to the introduced lesion. Gas exchange data showed that in the absence of PEPC, Rubisco appeared to be fixing CO2 directly, while plasmodesmatal density was increased in the low-PEPC plants, potentially as a compensation mechanism to allow higher CO2 flux into BSCs. This also represents a route for increased CO2 leakage from BSCs, which would normally be recycled by PEPC in the mesophyll. Investigation of the signalling pathways involved in this response represents a challenging next step in determining how gbs is modulated. (Summary by Mike Page) Plant Physiol. 10.1104/pp.18.00618
Receptor kinase THESEUS1 is a RALF 34 receptor with roles in lateral root development
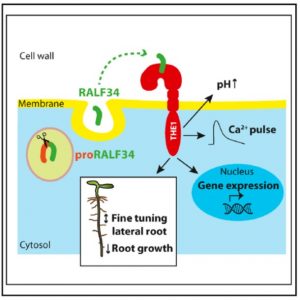 RALF peptides were identified nearly 20 years ago, as small peptides that induce rapid alkalinisation of the culture medium when added to cell suspension cultures. Previously, members of the receptor-like kinase (RLK) family including FERONIA (FER) have been identified as RALF receptors. A related RLK, THESEUS1 (THE1), was known to have a role in cell wall sensing and responses, but its ligand was not known. Now, Gonneau and Desprez et al. have shown that RALF34 is a ligand for THE1. The biological relevance of this interaction was demonstrated by several methods including genetics, co-occuring expression domains, and downstream gene expression studies, as well as micro-scale thermophoresis (MST) binding assays. The authors also demonstrate an important role for the THE1/RALF34 interaction in lateral root initiation. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2018.05.075
RALF peptides were identified nearly 20 years ago, as small peptides that induce rapid alkalinisation of the culture medium when added to cell suspension cultures. Previously, members of the receptor-like kinase (RLK) family including FERONIA (FER) have been identified as RALF receptors. A related RLK, THESEUS1 (THE1), was known to have a role in cell wall sensing and responses, but its ligand was not known. Now, Gonneau and Desprez et al. have shown that RALF34 is a ligand for THE1. The biological relevance of this interaction was demonstrated by several methods including genetics, co-occuring expression domains, and downstream gene expression studies, as well as micro-scale thermophoresis (MST) binding assays. The authors also demonstrate an important role for the THE1/RALF34 interaction in lateral root initiation. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2018.05.075
What a difference a base makes: A single nucleotide confers Alternaria resistance in apple ($)
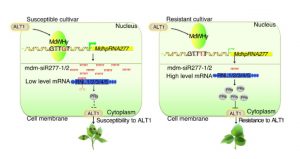 Golden Delicious apples are particularly susceptible to the fungal pathogen Alternaria alternaria f. sp. mali. Zhang et al. have traced this susceptibility to a single nucleotide in the promoter of gene encoding a hairpin RNA (hpRNA), MdhpRNA277. This hpRNA produces small RNAs that selectivey target five resistance genes. In susceptible plants, the hpRNA is produced and R genes silenced. In resistant plants, the single-nucleotide polymorphism disrupts binding of a transcription factor and so expression of the hpRNA, allowing high-level expression of the targeted R genes, and resistance to the fungus. As the authors report, “The ability to identify apple lines that are resistant to this devastating disease makes this SNP a valuable tool for the apple breeding industry.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00042
Golden Delicious apples are particularly susceptible to the fungal pathogen Alternaria alternaria f. sp. mali. Zhang et al. have traced this susceptibility to a single nucleotide in the promoter of gene encoding a hairpin RNA (hpRNA), MdhpRNA277. This hpRNA produces small RNAs that selectivey target five resistance genes. In susceptible plants, the hpRNA is produced and R genes silenced. In resistant plants, the single-nucleotide polymorphism disrupts binding of a transcription factor and so expression of the hpRNA, allowing high-level expression of the targeted R genes, and resistance to the fungus. As the authors report, “The ability to identify apple lines that are resistant to this devastating disease makes this SNP a valuable tool for the apple breeding industry.” (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00042
Phytophthora effector exploits host susceptibility factor NRL1
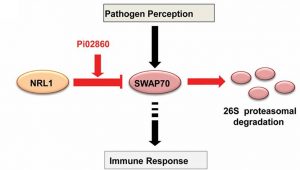 Some pathogen effectors disarm a plant’s immunity directly, for example through targeting resistance genes, whereas others work indirectly, through the plants own susceptibility (S) factors. He et al. provide an example of the latter. Previously, the authors showed that the Phytophthora infestans effector Pi02860 interacts with NRL1, a host S factor that is required for host susceptibility. NRL1 is a substrate adaptor component of a CULLIN3-associated ubiquitin E3 ligase. In this new work, they show that the presence of the effector enhances the interaction between NRL1 and SWAP70, a positive regulator of immunity, leading to SWAP70s degradation. As the authors observe, their research shows that “preventing the degradation of SWAP70 … provides a strategy to combat potato late blight.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1808585115
Some pathogen effectors disarm a plant’s immunity directly, for example through targeting resistance genes, whereas others work indirectly, through the plants own susceptibility (S) factors. He et al. provide an example of the latter. Previously, the authors showed that the Phytophthora infestans effector Pi02860 interacts with NRL1, a host S factor that is required for host susceptibility. NRL1 is a substrate adaptor component of a CULLIN3-associated ubiquitin E3 ligase. In this new work, they show that the presence of the effector enhances the interaction between NRL1 and SWAP70, a positive regulator of immunity, leading to SWAP70s degradation. As the authors observe, their research shows that “preventing the degradation of SWAP70 … provides a strategy to combat potato late blight.” (Summary by Mary Williams) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1808585115
Best of both worlds: a free living insect with an endophytic-like feeding strategy
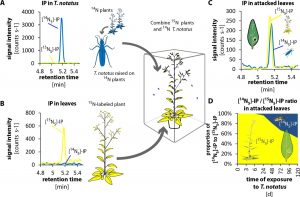 Endophytic insects, which spend much of their life inside of plant tissues (e.g., leaf miners and gall-forming insects) are well-known for manipulating plant host physiology to their advantage, unlike free living herbivore insects whose game plan relies on their ability to move and find the most nutritious part of the plant or the less defended one. However, in this article Brütting et al. report that when the free living mirid (a large family of sap-sucking bugs) Tupiocoris notatus is feeding on Nicotiana attenuata leaves, nutrient levels remain stable and cytokinin levels rise despite tissue damage produced by the insect. Through 15N-labeling experiments the authors find that the mirid transfers IP (a form of cytokinin) to its host plant most likely through salivary secretion. Increasing the amount of cytokinin in the leaf changes the source/sink relationship relocating nutrients for the insect to feed on. This opens the possibility of finding other similar cases that combine both lifestyles and shows the complexity of plant-herbivore interactions. (Summary by Mariana Antonietti) eLIFE 10.7554/eLife.36268.001
Endophytic insects, which spend much of their life inside of plant tissues (e.g., leaf miners and gall-forming insects) are well-known for manipulating plant host physiology to their advantage, unlike free living herbivore insects whose game plan relies on their ability to move and find the most nutritious part of the plant or the less defended one. However, in this article Brütting et al. report that when the free living mirid (a large family of sap-sucking bugs) Tupiocoris notatus is feeding on Nicotiana attenuata leaves, nutrient levels remain stable and cytokinin levels rise despite tissue damage produced by the insect. Through 15N-labeling experiments the authors find that the mirid transfers IP (a form of cytokinin) to its host plant most likely through salivary secretion. Increasing the amount of cytokinin in the leaf changes the source/sink relationship relocating nutrients for the insect to feed on. This opens the possibility of finding other similar cases that combine both lifestyles and shows the complexity of plant-herbivore interactions. (Summary by Mariana Antonietti) eLIFE 10.7554/eLife.36268.001
Carbon storage and land-use strategies in agricultural landscapes across three continents
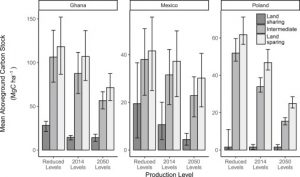 As we face increasingly odd weather patterns resulting from elevated CO2 emissions to the atmosphere, the question of how best to balance the need to produce food with the desire to minimize CO2 emissions becomes increasingly urgent. Williams et al. compared the effects of different types of land-use strategies on above-ground carbon stocks. They found that any agricultural use led to a decrease in carbon storage as compared to natural habitats. When they compared three different land-use strategies (land sharing with low-yield agriculture, land sparing with high-yield agriculture, and an intermediate strategy), the highest above-ground carbon stocks were maintained with the high-yield, land sparing strategy. Although they conclude that setting aside natural habitats and employing high-yield methods is the most beneficial for carbon storage, they recognize that “land sparing is unlikely to occur passively” but that it needs to involve land-use zoning, taxes or subsidies, and strategic investment to alter the relative profitability of agriculture. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2018.05.087
As we face increasingly odd weather patterns resulting from elevated CO2 emissions to the atmosphere, the question of how best to balance the need to produce food with the desire to minimize CO2 emissions becomes increasingly urgent. Williams et al. compared the effects of different types of land-use strategies on above-ground carbon stocks. They found that any agricultural use led to a decrease in carbon storage as compared to natural habitats. When they compared three different land-use strategies (land sharing with low-yield agriculture, land sparing with high-yield agriculture, and an intermediate strategy), the highest above-ground carbon stocks were maintained with the high-yield, land sparing strategy. Although they conclude that setting aside natural habitats and employing high-yield methods is the most beneficial for carbon storage, they recognize that “land sparing is unlikely to occur passively” but that it needs to involve land-use zoning, taxes or subsidies, and strategic investment to alter the relative profitability of agriculture. (Summary by Mary Williams) Curr. Biol. 10.1016/j.cub.2018.05.087



