Unraveling the epitranscriptomic-chromatin axis: How NERD regulates flowering in Arabidopsis
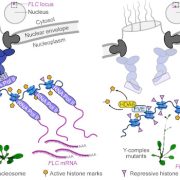
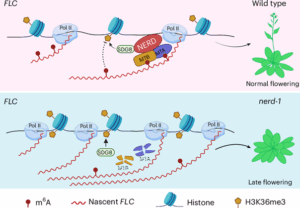 Flowering in Arabidopsis thaliana is governed by complex genetic and epigenetic networks. A central component of this regulation is the floral repressor FLOWERING LOCUS C (FLC), whose expression is modulated by histone modifications, particularly the activating H3K36me3 and the repressive H3K27me3 marks. While these chromatin-level mechanisms have been extensively studied, emerging evidence highlights a crucial role of RNA modifications, collectively termed epitranscriptomics, in fine-tuning gene expression. A recent study by Shao et al. uncovers a pivotal function for the plant-specific CCCH-type zinc finger protein NERD (NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION) in modulating N6-methyladenosine (m⁶A) deposition on FLC transcripts. The authors show that NERD physically interacts with core components of the m⁶A methyltransferase complex (MTA and MTB) to promote m⁶A modification of nascent RNA. This N6-methyladenosine (m⁶A) deposition is known to act as a transcriptional repressor. Through mass spectrometry and m⁶A RNA immunoprecipitation sequencing, the study reveals that nerd-1 mutants exhibit globally reduced m⁶A levels in both nuclear and mature mRNAs. Intriguingly, NERD interacts with the H3K36me3 methyltransferase SET DOMAIN GROUP 8 (SDG8), establishing a mechanistic link between RNA methylation and chromatin regulation. Loss of NERD function leads to increased levels of H3K36me3 at FLC, correlating with elevated transcription of this floral repressor and delayed flowering. This study uncovers a novel epitranscriptomic–chromatin regulatory axis in flowering control, highlighting the intricate cross-talk between RNA and chromatin landscapes and opening new avenues for exploring RNA-based gene regulations in plant development. (Summary by Gourav Arora @gouravarora.bsky.social) Nature Plants 10.1038/s41477-025-01945-7
Flowering in Arabidopsis thaliana is governed by complex genetic and epigenetic networks. A central component of this regulation is the floral repressor FLOWERING LOCUS C (FLC), whose expression is modulated by histone modifications, particularly the activating H3K36me3 and the repressive H3K27me3 marks. While these chromatin-level mechanisms have been extensively studied, emerging evidence highlights a crucial role of RNA modifications, collectively termed epitranscriptomics, in fine-tuning gene expression. A recent study by Shao et al. uncovers a pivotal function for the plant-specific CCCH-type zinc finger protein NERD (NEEDED FOR RDR2-INDEPENDENT DNA METHYLATION) in modulating N6-methyladenosine (m⁶A) deposition on FLC transcripts. The authors show that NERD physically interacts with core components of the m⁶A methyltransferase complex (MTA and MTB) to promote m⁶A modification of nascent RNA. This N6-methyladenosine (m⁶A) deposition is known to act as a transcriptional repressor. Through mass spectrometry and m⁶A RNA immunoprecipitation sequencing, the study reveals that nerd-1 mutants exhibit globally reduced m⁶A levels in both nuclear and mature mRNAs. Intriguingly, NERD interacts with the H3K36me3 methyltransferase SET DOMAIN GROUP 8 (SDG8), establishing a mechanistic link between RNA methylation and chromatin regulation. Loss of NERD function leads to increased levels of H3K36me3 at FLC, correlating with elevated transcription of this floral repressor and delayed flowering. This study uncovers a novel epitranscriptomic–chromatin regulatory axis in flowering control, highlighting the intricate cross-talk between RNA and chromatin landscapes and opening new avenues for exploring RNA-based gene regulations in plant development. (Summary by Gourav Arora @gouravarora.bsky.social) Nature Plants 10.1038/s41477-025-01945-7