Uncovering new start sites for plant viral protein synthesis
Chiu et al. characterize hidden viral genes in tomato yellow leaf curl Thailand virus.
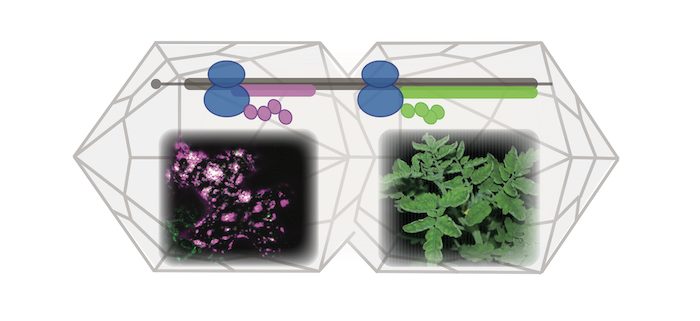
Background: When viruses such as begomoviruses infect plants, they hijack the host translation machinery to drive the expression of their proteins. This process causes viral disease, leading to great yield losses in crops such as tomato. To develop effective antiviral strategies, pathogenesis studies are needed to investigate how virus-encoded proteins contribute to the viral lifecycle and disease symptom development.
Question: Plant viruses with densely packed genomes employ non-canonical translational strategies to increase their coding capacity. However, the diverse translational strategies used make it challenging to define the full set of viral genes.
Findings: To obtain a global view of viral genes expressed during the lifecycle of tomato yellow leaf curl Thailand virus, we investigated the sites on viral mRNAs that can initiate protein translation using a technique for globally mapping in vivo translating ribosomes. We found multiple initiation sites that encode viral factors important for disease symptom development in tomato. Our findings suggest that the complexity and diversity of plant virus coding capacity are underappreciated.
Next steps: Our findings raise the next grand questions: How are the newly identified open reading frames induced during viral infection, and how do they contribute to viral pathogenesis? Thus, the next important steps are to elucidate the mechanistic basis of the cis- and trans-regulatory factors involved in viral gene expression and how these new viral factors coordinate the viral lifecycle and function during pathogenesis in plants.
Ching-Wen Chiu, Ya-Ru Li, Cheng-Yuan Lin, Hsin-Hung Yeh, and Ming-Jung Liu (2022). Translation initiation landscape profiling reveals hidden open reading frames required for the pathogenesis of tomato yellow leaf curl Thailand virus. https://doi.org/10.1093/plcell/koac019