Ubiquitous ubiquitin: The K63 ubiquitinome
Polyubiquitination, the sequential attachment of the small 8kDa globular protein ubiquitin (Ub) to target proteins, represents a major post-translational modification that ultimately determines the substrate’s cellular fate. Ub conjugation is an extremely versatile form of protein regulation since any of the Ub’s seven lysine residues can form such polymeric chains, plus the process itself is reversible. Historically, polyubiquitination of substrates has been associated with their proteasomal degradation, but it is mainly Lys48 (UbK48) linkages that are responsible for such targeting. Much less is known about the functions of the remaining UbK linkages in plants, although UbK63 chains have been associated with endocytosis and DNA damage tolerance in yeast and mammalian systems (reviewed in Mukhopadhyay and Riezman, 2007).
 Here, Romero-Barrios et al. (2020) employ both large-scale protein-protein interaction (PPI) and sensor-based proteomic techniques to define UbK63 polyubiquitination networks in Arabidopsis thaliana (Arabidopsis) and shed light on the molecular machinery driving this modification. Ubiquitination is a three-stage process whereby Ub is activated by an E1 enzyme, transferred to an E2 ubiquitin-conjugating enzyme, and attached to a specific substrate recruited via an E3 ubiquitin ligase. In humans and yeast, UbK63 polyubiquitination is mediated by UBIQUITIN CONJUGATING ENZYME13 (UBC13)/UEV1 E2s in concert with one of several hundred possible E3 ligases or by a relay mechanism in which ubiquitin is passed from E2s to HECT E3s before its ligation to a substrate. Plant genomes, however, typically encode very few HECT E3s, but they do encode several UBC13/UEV1 proteins, including UBC35 and 36 in Arabidopsis. Functional analysis of a double ubc35 ubc36 knockout mutant, which exhibited severe growth and developmental defects, alongside an inducible ubc35 ubc36 knockdown line, revealed a major reduction in cellular abundance of UbK63 polyubiquitinated proteins together with significant transcriptional reprogramming. These results strongly implicate UBC35 and 36 E2s as the key mediators of UbK63 modifications in plants.
Here, Romero-Barrios et al. (2020) employ both large-scale protein-protein interaction (PPI) and sensor-based proteomic techniques to define UbK63 polyubiquitination networks in Arabidopsis thaliana (Arabidopsis) and shed light on the molecular machinery driving this modification. Ubiquitination is a three-stage process whereby Ub is activated by an E1 enzyme, transferred to an E2 ubiquitin-conjugating enzyme, and attached to a specific substrate recruited via an E3 ubiquitin ligase. In humans and yeast, UbK63 polyubiquitination is mediated by UBIQUITIN CONJUGATING ENZYME13 (UBC13)/UEV1 E2s in concert with one of several hundred possible E3 ligases or by a relay mechanism in which ubiquitin is passed from E2s to HECT E3s before its ligation to a substrate. Plant genomes, however, typically encode very few HECT E3s, but they do encode several UBC13/UEV1 proteins, including UBC35 and 36 in Arabidopsis. Functional analysis of a double ubc35 ubc36 knockout mutant, which exhibited severe growth and developmental defects, alongside an inducible ubc35 ubc36 knockdown line, revealed a major reduction in cellular abundance of UbK63 polyubiquitinated proteins together with significant transcriptional reprogramming. These results strongly implicate UBC35 and 36 E2s as the key mediators of UbK63 modifications in plants.
To identify which of the ~1500 Arabidopsis E3 ligases interact with UBC35/36 to facilitate UbK63 chain formation, the authors utilized a high coverage yeast two-hybrid (Y2H) approach. Only 13 (mainly RING/U-box family) E3s were detected in the screen, attesting to the existence of E2-E3 interaction specificity. By widening this UBC35/36-E3 PPI network further to include interactors of these 13 E3 ligases, the authors also identified several putative UbK63 substrates, generating an extensive UbK63 polyubiquitination interactome network.
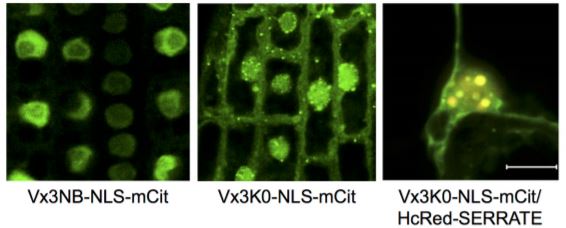
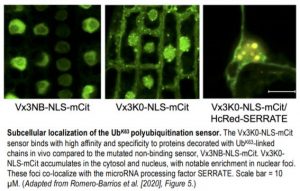
However, since the E2s and not the E3s determine Ub-linkage preference, binary PPIs between E3 ligases and target proteins may potentially represent non-UbK63 polyubiquitination events. To validate and extend the UbK63-specific polyubiquitination network in planta, the authors modified their previously described in vivo sensor for UbK63 polyubiquitination (Vx3K0-GFP) (Johnson and Vert, 2016) with a nuclear-localization sequence (Vx3K0-NLS-mCitrine) to capture both cytosolic and nuclear targets. Proteins decorated with UbK63-linked chains were immunoprecipitated from stable Arabidopsis lines This in vivo proteomics approach identified nearly 400 proteins, which given the Vx3K0 sensor’s strong selectivity for UbK63 over UbK48 (Johnson and Vert, 2016), represent high-confidence UBC35/36 targets.
Although, somewhat surprisingly, limited overlap was observed in the membership of proteins identified by the transcriptomic, sensor-based proteomic, and interactome analyses, taken together, a global picture emerges of the overall contribution of the putative UbK63 ubiquitinome to cellular functioning. In agreement with previous studies in both plants and yeast/mammalian systems, UbK63 polyubiquitination is associated with vesicular trafficking, membrane transport, and developmental processes. However, the use of the nuclear-localized UbK63 sensor also revealed that several nuclear functions involving various aspects of RNA processing required UbK63 polyubiquitination, alongside multiple components of the nuclear import machinery and specific histones associated with DNA damage responses.
After proteasome-targeting UbK48 polyubiquitination, UbK63 chains represent the most abundant linkage type, but the importance of this modification in plants has only recently started to be appreciated. This study by Romero-Barrios et al. (2020) highlights the breadth of growth and developmental processes regulated by UbK63 polyubiquitination and identifies the two major E2 Ub-conjugating enzymes responsible for this linkage type. Further characterization of Ub signaling networks underpinning plant growth and development will require further insight into the functional significance of the remaining UbK-linked chains as well as the proteins responsible for linkage-type discrimination.
Emily Breeze
School of Life Sciences, University of Warwick, Coventry, UK
ORCID ID: 0000-0001-5383-5448
REFERENCES
Johnson, A. and Vert, G. (2016). Unraveling K63 Polyubiquitination Networks by Sensor-Based Proteomics. Plant Physiol. 171: 1808–1820.
Mukhopadhyay, D. and Riezman, H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205.
Romero-Barrios, N., Monachello, D., Dolde, U., Wong, A., Clemente, H.S., Cayrel, A., Johnson, A., Lurin, C. and Vert, G. (2020) Advanced cataloging of K63 polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. Plant Cell 32: doi:10.1015/tpc19.00568