Truncated organellar C-to-U RNA editing enzyme requires active site complementation
By Sachiko Toma-Fukai1,2 Mizuki Takenaka3 and Toshiyuki Shimizu1
1 Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
2 Present address: Graduate School of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama-cho, Ikoma, Nara 630-0192, Japan
3 Department of Botany, Graduate School of Science, Kyoto University, Kyoto, Japan.
Background: In terrestrial plants, C-to-U RNA editing occurs in mitochondrial and chloroplast genes. More than one hundred pentatricopeptide repeat (PPR) proteins have been identified as specificity factors for RNA editing. The DYW deaminase domain in PPR proteins serves as an enzyme for C-to-U conversion. More than half of the PPR-type RNA editing factors, however, lack the DYW cytidine deaminase domain. Genetic analyses have suggested that their enzymatic activity arises by association with a family of DYW1-like proteins that contain an N-terminally truncated DYW domain. Nevertheless, the mechanism underlying the activation of the truncated DYW deaminase remains largely unknown.
Question: What is the structural mechanism underlying activation of the DYW1-like deaminase?
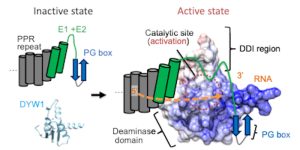 Findings: We determined the crystal structure of the N-terminally truncated DYW domain from Arabidopsis thaliana DYW1. DYW1 forms a cytidine deaminase fold and a C-terminal DYW motif harboring two zinc ions but lacks two structurally conserved β-strands corresponding to the PG box. The recruitment of the PG box and its flanking region from CRR4 likely enables the pocket to accommodate the substrate, conferring catalytic activity to DYW1. In vivo RNA editing assays corroborate that the presence of a PG box in the vicinity of DYW1 is required for its deaminase activity.
Findings: We determined the crystal structure of the N-terminally truncated DYW domain from Arabidopsis thaliana DYW1. DYW1 forms a cytidine deaminase fold and a C-terminal DYW motif harboring two zinc ions but lacks two structurally conserved β-strands corresponding to the PG box. The recruitment of the PG box and its flanking region from CRR4 likely enables the pocket to accommodate the substrate, conferring catalytic activity to DYW1. In vivo RNA editing assays corroborate that the presence of a PG box in the vicinity of DYW1 is required for its deaminase activity.
Next steps: The results described here will provide valuable information for developing the DYW domain as a controllable genetic manipulation tool. Further structural studies of DYW1 in complex with its partner proteins are required to shed light on its editing mechanism.
Sachiko Toma-Fukai, Yuto Sawada, Ayako Maeda, Hikaru Shimizu, Toshiharu Shikanai, Mizuki Takenaka and Toshiyuki Shimizu. (2023). Structural insight into the activation of an Arabidopsis organellar C-to-U RNA editing enzyme by active site complementation https://doi.org/10.1093/plcell/koac318