Translational Recoding Corrects Frameshift Mutations in Chloroplasts
Malinova et al. provide genetic and biochemical evidence for ribosomal frameshifting in chloroplasts. Plant Cell https://doi.org/10.1093/plcell/koab050
By Irina Malinova and Stephan Greiner
Background: Translational recoding or ribosomal frameshifting – a process when ribosomes either omit nucleotide(s) or move back along the mRNA template during protein synthesis – is typical for the bacteria. Chloroplasts (photosynthetic plant organelles) possess their own genome that evolutionary derives from cyanobacteria. However, no clear evidence of ribosomal frameshifting operating in chloroplasts has been published so far.
Question: We addressed whether ribosomal frameshifting functions in the chloroplasts, and if so, what role it plays in plants.
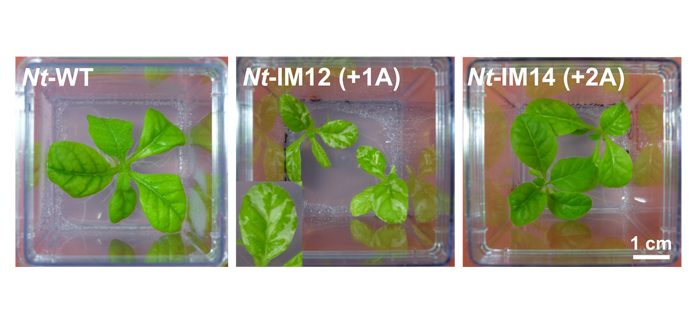
Findings: The spontaneous chloroplast mutant I-iota of the evening primrose (Oenothera) displays mottled leaves. The underlying mutation is a single adenine insertion in oligoA stretch very early in the atpB gene. The latter encodes the beta subunit of chloroplast ATP synthase. According to the standard rules of genetic coding, this mutation should lead to the formation of a dysfunctional protein. However, a full-length AtpB protein was detected in I-iota leaves. This indicates that a correction mechanism is acting in the mutant. To analyze this phenomenon in more detail, we generated a series of tobacco chloroplast mutants. Interestingly, an introduction of the I-iota mutation in tobacco [Nt-IM12(+1A)] resulted in phenotype similar to that of the I-iota plants. Additionally, synthesis of functional AtpB protein was confirmed. By contrast, tobacco mutants with an insertion of two adenines within the oligoA stretch of atpB [Nt-IM14 (+2A)] were green and could even grow on soil. These results and further analyses of ATP synthase and mRNA confirmed that Nt-IM14 (+2A) can efficiently correct the +2A mutation via ribosomal frameshifting. Strikingly, it enables restoration of photoautotrophic growth.
Next steps: Besides correction of mutations, we aim to identify to what extent ribosomal frameshifting is a mechanism of gene regulation in the chloroplast.
Irina Malinova, Arkadiusz Zupok, Amid Massouh, Mark Aurel Schöttler, Etienne H. Meyer, Liliya Yaneva-Roder, Witold Szymanski, Margit Rößner, Stephanie Ruf, Ralph Bock, Stephan Greiner. (2021). Correction of Frameshift Mutations in the atpB Gene by Translational Recoding in Chloroplasts of Oenothera and Tobacco. Plant Cell. https://doi.org/10.1093/plcell/koab050