Too Close to the Flame: Duplicated ICARUS Genes and Growth at Higher Temperatures
Plants can adjust their growth and reproduction cycles according to the local climate they experience, but how will they respond when ambient temperatures rise? Most Arabidopsis accessions respond by increasing cell elongation and accelerated flowering, but some cannot cope with higher temperatures and stop growing.
Such was the premise that led to the cloning of ICARUS1 (ICAR1) back in 2015. ICA1 was mapped in two F2 populations: Don-0 x Ler and Sij-4 x Col-0; both Don-0 and Sij-4 displayed severe growth defects at 28ºC, including the loss of high-temperature-induced hypocotyl and petiole elongation, higher DNA content and endoreduplication, as well as delayed flowering (Zhu et al., 2015). The two resulting mapping intervals overlapped only over a ~6 kbp region containing a single annotated gene: ICA1, encoding a tRNAHis guanylyl transferase important for tRNA maturation, translation, and DNA replication. T-DNA insertion alleles and amiRNA (artificial microRNA) lines in the Col-0 background recapitulated the growth phenotypes at 28ºC; conversely, the Don-0 phenotype was rescued by introduction of the ICA1 allele from Ler. Together, these results strongly argued that ICA1 was the causal locus.
Now, Méndez-Vigo and co-authors revisit the role of ICA1 in high temperature responses: the phenotype observed in Sij-4 and Don-0 is actually caused by the interaction of ICA1 and its closely linked duplicate, ICA2. Indeed, one nagging and unresolved point from the previous study was the size of the mapping interval in Don-0 x Ler: almost 400 kbp, approximately 10-times that of the Sij-4 x Col-0 interval, pointing to a more complex picture. The genetics truly make sense only when invoking a second linked locus that interacts with ICA1: ICA2, a homolog of ICA1 located ~ 300 kbp away.
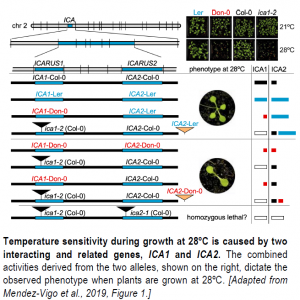 How does a two-gene model help explain the observed temperature sensitivity seen in Don-0? It does so by considering an allelic series encoded by ICA1 and ICA2 across accessions (see Figure). From the 2015 study, we knew that ICA1-Col-0 and ICA1-Ler were active, and ICA1-Don-0 much less so. Here, the authors show that the ICA2 alleles ICA2-Col-0 and ICA2-Don-0 only have weak activity; in addition, a T-DNA insertion in ICA2 in Col-0 only has very mild phenotypes. In agreement with this notion, ICA2-Don-0 cannot rescue the temperature-sensitivity phenotype of ica1 T-DNA insertions in Col-0, although ICA2-Ler can, indicating that ICA2-Ler is more active and a dominant allele. This dominance was likely responsible for the much larger mapping interval in Don-0 x Ler, as temperature sensitivity only became apparent when both ICA1 and ICA2 came from Don-0 (Zhu et al., 2015; Méndez-Vigo et al., 2019). The situation is much simpler in the Don-0 x Col-0 mapping population, where only ICA1 matters in this case. Yet, ICA2-Don-0 must have some activity to compensate for the loss of function from ICA1-Don-0, since ica1 ica2 double mutants could not be isolated.
How does a two-gene model help explain the observed temperature sensitivity seen in Don-0? It does so by considering an allelic series encoded by ICA1 and ICA2 across accessions (see Figure). From the 2015 study, we knew that ICA1-Col-0 and ICA1-Ler were active, and ICA1-Don-0 much less so. Here, the authors show that the ICA2 alleles ICA2-Col-0 and ICA2-Don-0 only have weak activity; in addition, a T-DNA insertion in ICA2 in Col-0 only has very mild phenotypes. In agreement with this notion, ICA2-Don-0 cannot rescue the temperature-sensitivity phenotype of ica1 T-DNA insertions in Col-0, although ICA2-Ler can, indicating that ICA2-Ler is more active and a dominant allele. This dominance was likely responsible for the much larger mapping interval in Don-0 x Ler, as temperature sensitivity only became apparent when both ICA1 and ICA2 came from Don-0 (Zhu et al., 2015; Méndez-Vigo et al., 2019). The situation is much simpler in the Don-0 x Col-0 mapping population, where only ICA1 matters in this case. Yet, ICA2-Don-0 must have some activity to compensate for the loss of function from ICA1-Don-0, since ica1 ica2 double mutants could not be isolated.
Most duplicated genes tend to diverge in function over time, but here both copies remain functional and participate in plant tolerance to higher temperatures, although the precise mechanism remains unclear. A possible role in adaptation to climate variables is also hinted at by the geographic range of the minor-allele ICA2-Ler, restricted to accessions from northern Europe and Asia. If decreasing ICA1/ICA2 activity results in temperature sensitivity, perhaps raising it by introducing ICA2-Ler in other accessions or crops might prove useful in climate tolerance.
References:
Méndez-Vigo B, Ausín I, Zhu W, Mollá-Morales A, Balasubramanian S and Alonso-Blanco C (2019). Genetic interaction and molecular evolution of the duplicated genes ICARUS2 and ICARUS1 confer growth adaptation to ambient temperature in Arabidopsis. The Plant Cell. Published April 2019. DOI: https://doi.org/10.1105/tpc.18.00938
Zhu W, Ausin I, Seleznev A, Mendez-Vigo B, Pico F.X, Sureshkumar S, Sundaramoorthi V, Bulach D, Powell D, Seemann T, Alonso-Blanco C, and Balasubramanian, S (2015). Natural variation identifies ICARUS1, a universal gene required for cell proliferation and growth at high temperatures in Arabidopsis thaliana. PLoS Genet. 11, e1005085.



