The R-loop: An Additional Chromatin Feature for Gene Regulation in Arabidopsis
R-loops are three-stranded nucleic acid structures composed of a DNA-RNA hybrid and a displaced single-stranded DNA. R-loops are stable structures as they form an intermediate A/B conformation of the RNA-DNA hybrid that is more stable than the B form of dsDNA or A form of the dsRNA. Recent evidence reveals the occurrence of R-loops in multiple organisms from prokaryotes to eukaryotes, and their essential roles in transcription, chromatin modification, DNA damage response, and genome stability. It is also speculated that R-loops can lead to DNA damage and genome instability as a result of the exposure of ssDNA, making them susceptible to lesions and transcription-associated mutagenesis and recombination. In human cell lines, C. elegans, and S. cerevisiae, R-loop accumulation is linked to chromatin compaction leading to replication fork stalling, transcription-replication collisions, and double-strand breaks (Skourti-Stathaki and Proudfoot, 2014). Arabidopsis R-loops are characterized by GC and AT skews, strong enrichment in promoters and gene bodies, association with repetitive elements and noncoding RNA, and negative correlation with CG hypermethylation (Xu et al., 2017). Several patterns of R-loops in Arabidopsis are distinct from those in mammals, such as accumulation of antisense R-loops, low levels of R-loops in gene terminators, and strong association with targets of the plant-specific RNA-directed DNA methylation pathway.
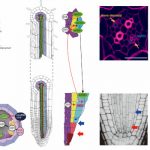
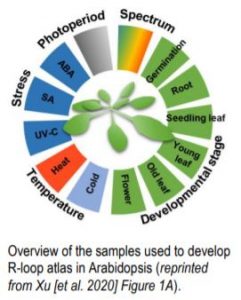 Functional studies of R-loops in plants have been limited to specific R-loop loci (Santos-Pereira and Aguilera, 2015). In new work, Xu et al. (2020) set out to profile genome-wide R-loop dynamics in Arabidopsis over several developmental stages and in response to different biotic and abiotic stressors (Figure). Thus, they developed the first R-loop atlas of land plant development and response to environmental stimuli, greatly expanding our understanding of R-loop distribution patterns in Arabidopsis. The authors used a single-strand DNA ligation-based library constructed from DNA:RNA hybrid immunoprecipitation followed by next-generation sequencing(ssDRIP-seq) to investigate R-loop dynamics. They found that the R-loop distribution patterns were relatively unchanged during most of the tested conditions except during phase switches such as flowering, germination, and recovery from stress treatments. Global changes in R-loops during plant transition from vegetative to reproductive phases were restored to juvenile patterns during seed germination. R-loops in the sense orientation was highly enriched on the gene bodies (sGB-R-loops) and antisense R loops were enriched around the transcription start sites (asTSS-R-loops). The authors also used immunostaining with S9.6 antibodies that detect DNA:RNA hybrids, to show a decrease in R-loop levels during flowering and germination, consistent with their sequencing data.
Functional studies of R-loops in plants have been limited to specific R-loop loci (Santos-Pereira and Aguilera, 2015). In new work, Xu et al. (2020) set out to profile genome-wide R-loop dynamics in Arabidopsis over several developmental stages and in response to different biotic and abiotic stressors (Figure). Thus, they developed the first R-loop atlas of land plant development and response to environmental stimuli, greatly expanding our understanding of R-loop distribution patterns in Arabidopsis. The authors used a single-strand DNA ligation-based library constructed from DNA:RNA hybrid immunoprecipitation followed by next-generation sequencing(ssDRIP-seq) to investigate R-loop dynamics. They found that the R-loop distribution patterns were relatively unchanged during most of the tested conditions except during phase switches such as flowering, germination, and recovery from stress treatments. Global changes in R-loops during plant transition from vegetative to reproductive phases were restored to juvenile patterns during seed germination. R-loops in the sense orientation was highly enriched on the gene bodies (sGB-R-loops) and antisense R loops were enriched around the transcription start sites (asTSS-R-loops). The authors also used immunostaining with S9.6 antibodies that detect DNA:RNA hybrids, to show a decrease in R-loop levels during flowering and germination, consistent with their sequencing data.
ONSEN, a subgroup of copia-type retrotransposon activated during heat stress, showed a greater than 100-fold increase in sGB-R-loops after prolonged heat stress (LHS), and showed decreased levels of R-loops after recovery. Such associated changes in R-loop dynamics and RNA levels were only seen at the ONSEN loci and not in other heat stress-responsive gene loci or transposable elements. Analysis of RNA-seq data from prolonged heat stress and other published RNA-seq data from roots, flowers, leaves, and germinating seedlings showed a weak linear relationship for sGB-R-loops/asTSS-R-loops with RNA levels indicating weak coupling of global R-loop status with RNA levels. Interestingly, asTSS-R-loops were enriched in LHS differentially expressed genes suggesting their role in regulating transcriptionally dynamic genes. Furthermore, asTSS-R-loops were also significantly enriched with transcription factor binding sites on gene promoters. It is noteworthy that asTSS-R-loops have not been observed in other species potentially signifying their unique regulatory mechanism and function in Arabidopsis.
Research over the past decades in mammals has provided evidence that R-loops can be a source of genome instability and contribute to diseases in humans. It is not known if R-loops have such negative consequences for plant genomes. Xu et al. provide novel insights into the physiological roles and consequences of R-loops in Arabidopsis gene regulation during development and response to the environment. Future research is necessary to untangle the positive and negative effects of R-loops and improve our understanding of these key players in genome dynamics.
Sunil K. Kenchanmane Raju
Department of Plant Biology, Michigan State University.
ORCID ID: 0000-0001-8960-094X
REFERENCES
Skourti-Stathaki, K., & Proudfoot, N. J. (2014). A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28: 1384-1396.
Xu, W., Xu, H., Li, K., Fan, Y., Liu, Y., Yang, X., & Sun, Q. (2017). The R-loop is a common chromatin feature of the Arabidopsis genome. Nature Plants 3: 704-714.
Santos-Pereira, J. M., & Aguilera, A. (2015). R loops: new modulators of genome dynamics and function. Nature Rev. Genet. 16: 583-597.
Xu, W., Li, K., Li, S., Hou, Q., Zhang, Y., Liu, K., & Sun, Q. (2020). The R-loop Atlas of Arabidopsis Development and Responses to Environmental Stimuli. Plant Cell. DOI: https://doi.org/10.1105/tpc.19.00802.