Successive passaging of plant associated microbiome reveals robust habitat and host genotype-dependent selection (PNAS)
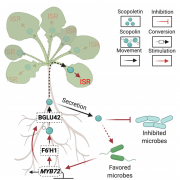
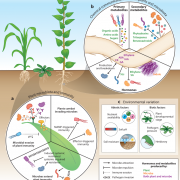
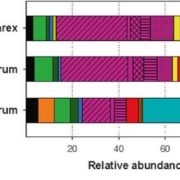
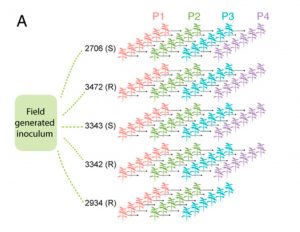 Study of the plant microbiome can contribute to sustainable agricultural systems through improvements to plant health and nutrition. One goal has been to select and design microbiome communities for specific functions. In a recent article, Morella et al. have used an experimental evolution approach to understand the robustness of the plant microbiome after colonization of the host. They inoculated five different genotypes of tomato with a diverse set of microbes, which were serially passaged four times from 45-day old plants. The phyllosphere bacterial and fungal communities were sequenced and analyzed after each passage, which revealed a loss of diversity and a loss of genotype effect. At the end of the first passage, 92% of initial inoculum operational taxonomic units (OTUs) were present, and the number reduced to 29% at the end of the fourth passage. Interestingly, the bacterial community from the end of the experiment was robust to invasion by the starting bacterial community. This indicates that efforts to support plant growth by inoculating them with a beneficial microbiome could be successful. (Summary by Mugdha Sabale) Proc. Natl. Acad. Sci. USA
Study of the plant microbiome can contribute to sustainable agricultural systems through improvements to plant health and nutrition. One goal has been to select and design microbiome communities for specific functions. In a recent article, Morella et al. have used an experimental evolution approach to understand the robustness of the plant microbiome after colonization of the host. They inoculated five different genotypes of tomato with a diverse set of microbes, which were serially passaged four times from 45-day old plants. The phyllosphere bacterial and fungal communities were sequenced and analyzed after each passage, which revealed a loss of diversity and a loss of genotype effect. At the end of the first passage, 92% of initial inoculum operational taxonomic units (OTUs) were present, and the number reduced to 29% at the end of the fourth passage. Interestingly, the bacterial community from the end of the experiment was robust to invasion by the starting bacterial community. This indicates that efforts to support plant growth by inoculating them with a beneficial microbiome could be successful. (Summary by Mugdha Sabale) Proc. Natl. Acad. Sci. USA