SoyOmics: A new tool to explore the transcriptome dynamics of soybean
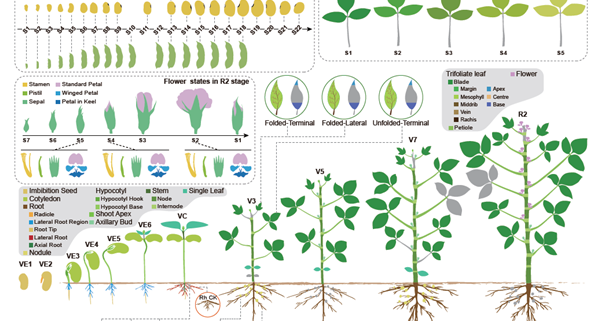
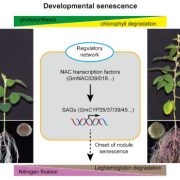
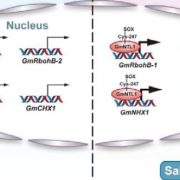
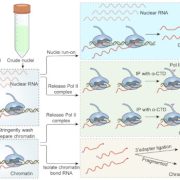
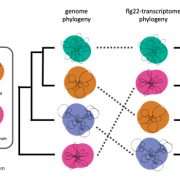
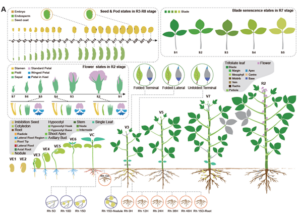 Soybean, originally domesticated in China, is widely recognized for its high protein and oil content, making it a crucial crop for human consumption, animal feed, and biofuel production. Despite its economic and agricultural significance, research tools and genomic resources for soybean have lagged behind those of other model species. The first genome assembly of a cultivated soybean variety was published 15 years ago, followed by the wild soybean genome nearly a decade later. While multi-omics data for soybean have grown rapidly in recent years, a centralized and user-friendly platform for accessing and analyzing these datasets remains underdeveloped. To bridge this gap, Fan and colleagues have established SoyOmics (https://ngdc.cncb.ac.cn/soyomics/transcriptome), a comprehensive database integrating bulk RNA-seq data from 314 samples spanning the soybean life cycle. Additionally, it includes single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq datasets from five key organs: root, nodule, shoot apical meristem, leaf, and stem. This platform provides an invaluable resource for the soybean research community, facilitating gene association mapping, network analysis, and functional studies to advance soybean biology and breeding efforts. (Summary by Ching Chan @ntnuchanlab) Molecular Plant 10.1016/j.molp.2025.02.003
Soybean, originally domesticated in China, is widely recognized for its high protein and oil content, making it a crucial crop for human consumption, animal feed, and biofuel production. Despite its economic and agricultural significance, research tools and genomic resources for soybean have lagged behind those of other model species. The first genome assembly of a cultivated soybean variety was published 15 years ago, followed by the wild soybean genome nearly a decade later. While multi-omics data for soybean have grown rapidly in recent years, a centralized and user-friendly platform for accessing and analyzing these datasets remains underdeveloped. To bridge this gap, Fan and colleagues have established SoyOmics (https://ngdc.cncb.ac.cn/soyomics/transcriptome), a comprehensive database integrating bulk RNA-seq data from 314 samples spanning the soybean life cycle. Additionally, it includes single-nucleus RNA sequencing (snRNA-seq) and Stereo-seq datasets from five key organs: root, nodule, shoot apical meristem, leaf, and stem. This platform provides an invaluable resource for the soybean research community, facilitating gene association mapping, network analysis, and functional studies to advance soybean biology and breeding efforts. (Summary by Ching Chan @ntnuchanlab) Molecular Plant 10.1016/j.molp.2025.02.003