Smashing barriers in biolistic plant transformation
A foundation of modern biotechnology is the ability to stably introduce foreign DNA into an organism. The two most widely used methods, Agrobacterium-mediated transformation and biolistics, are both steeped in a rich history of creative exploration into the molecular unknown. Agrobacterium research accelerated in the early 1970s, particularly with the discovery of the large Ti (tumor-inducing) plasmid of Agrobacterium that contained a region of transfer DNA (T-DNA). Culturing plant calli in autoclaved jelly jars, and long before the advent of PCR, Southern blots were first used to show that T-DNA fragments could stably integrate into the nuclear genome (Chilton et al., 1980; Chilton, 2001). On the other hand, the first manufactured biolistic “gene gun” was an actual gun; it shot a blank .22 caliber cartridge loaded with DNA-coated tungsten shards to integrate foreign DNA into the nuclear genome. While it has long been known that biolistic transformation violently integrates DNA in a largely random, unpredictable and imprecise way, the cellular mechanisms of damage repair and successful integration remain a complicated issue to disentangle.
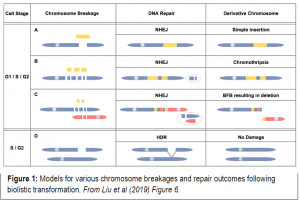 Liu et al. (2019) address the question of biolistic damage and repair in a controlled fashion. They biolistically transform three constructs into maize and rice callus, a linear 48 kb lambda phage, as well as two circular constructs, and assess the impact of genomic disruption and repair of successful transgenics using short-read Illumina whole genome resequencing, long-read PacBio sequencing and long-range Bionano optical mapping. After screening and resequencing transformed callus for the presence of lambda, they discover a multitude of different types and degrees of integration (Figure 1). Some of the transformants for both rice and maize contained one or just a few lambda insertions, with no other evidence of genome-wide damage such as deletions or rearrangements. These low-copy insertions were sometimes accompanied by small chromosomal deletions around the lambda insertion site. Oppositely, as shown in another rice transformant line λ-4, lambda was present in high copy and shattered into an array containing small pieces of genomic DNA derived from other parts of the genome.
Liu et al. (2019) address the question of biolistic damage and repair in a controlled fashion. They biolistically transform three constructs into maize and rice callus, a linear 48 kb lambda phage, as well as two circular constructs, and assess the impact of genomic disruption and repair of successful transgenics using short-read Illumina whole genome resequencing, long-read PacBio sequencing and long-range Bionano optical mapping. After screening and resequencing transformed callus for the presence of lambda, they discover a multitude of different types and degrees of integration (Figure 1). Some of the transformants for both rice and maize contained one or just a few lambda insertions, with no other evidence of genome-wide damage such as deletions or rearrangements. These low-copy insertions were sometimes accompanied by small chromosomal deletions around the lambda insertion site. Oppositely, as shown in another rice transformant line λ-4, lambda was present in high copy and shattered into an array containing small pieces of genomic DNA derived from other parts of the genome.
These findings opened up a major question: how exactly does a chromosome repair itself after a rather violent biolistic integration? Two known pathways are non-homologous end joining (NHEJ), where blunt-ended junctions fuse, and microhomology-mediated end joining (MMEJ), which uses small regions of aligned homology (longer than five nucleotides). Since both methods of repair lead to small deletions that flank the original break-site, the authors computationally detected these events and found evidence for both types, but NHEJ and MMEJ could not explain every detected case of successful lambda integration. There exists another major form of chromosomal repair, homology-directed repair (HDR), which can repair double-stranded DNA breaks using undamaged templates (such as sister chromatids) as guides. Indeed, rice line λ-4 and several other transformants showed evidence of HDR, where areas of the genome that had been damaged during transformation were seamlessly repaired.
Even more surprising, some cases of multiple simultaneous chromosome breakages may have created interchromosomal fusions, creating unstable dicentric chromosomes (with two centromeres) that segregate improperly to form trisomics. These unstable dicentric chromosomes can trigger the Breakage-Fusion-Bridge cycle to create even more downstream structural variation, famously described by Barbara McClintock (McClintock, 1938). In other cases, chromosome impact sites appear to have “shattered” and reassembled with copy number variations, rearrangements, and deletions in a phenomenon called chromothripsis.
What Liu et al. reiterates to us is that biolistic transformation creates a slew of diverse chromosome damage, which in turn triggers a variety of DNA integration and repair mechanisms. With the advent of precision Cas9- and Cpf-based editing tools, classic Agrobacterium-mediated transformation methods may remain superior over biolistic transformation if the goal is to minimize off-target genome damage.
REFERENCES
Chilton, M.-D. (2001). Agrobacterium. A Memoir. Plant Physiol. 125: 9–14.
Chilton, M.D., Saiki, R.K., Yadav, N., Gordon, M.P., and Quetier, F. (1980). T-DNA from Agrobacterium Ti plasmid is in the nuclear DNA fraction of crown gall tumor cells. Proc. Natl. Acad. Sci. U. S. A. 77: 4060–4064.
Liu, J., Nannas, N.J., Fu, F.-F., Shi, J., Aspinwall, B., Parrott, W.A., and Dawe, R.K. (2019). Genome-scale Sequence Disruption Following Biolistic Transformation in Rice and Maize. Plant Cell. https://doi.org/10.1105/tpc.18.00613
McClintock, B. (1938). The Production of Homozygous Deficient Tissues with Mutant Characteristics by Means of the Aberrant Mitotic Behavior of Ring-Shaped Chromosomes. Genetics 23: 315–376.



