RNA G-quadruplex structures exist and function in vivo (bioRxiv)
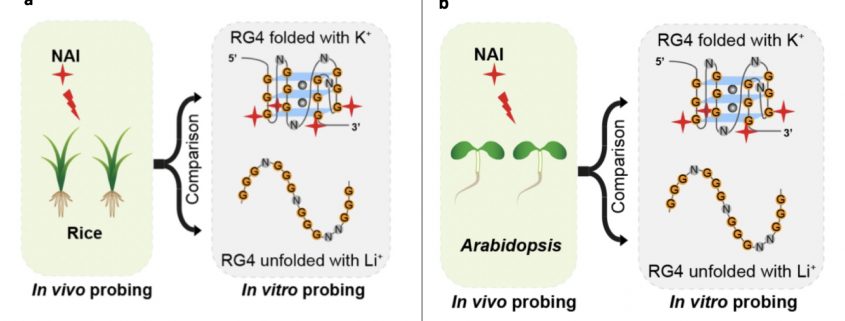
RNA G quadruplex (RG4) structures that include two or more layers of G-quartets can form in guanine-rich RNA sequences. These structures are known exist in vitro and are proposed to form in vivo as well. However, there is no direct evidence from plants showing the presence of these structures in vivo. Here, Yang et al. screened for potential RG4-forming sites and assessed them for their potential to fold as RG4 structures. In silico analysis suggested more than 60,000 potential RG4-forming sites (based on the presence of the GxLnGxLnGxLnGx sequence). RG4 structures cause reverse transcriptional stalling (RTS), and when this functional criterion (rG4-seq) was used fewer than 3000 potential RG4 sites were identified, mostly in coding sequences. The authors determined that the undetected sites are more likely to fold into stable secondary structures rather than RG4 structures. Sites identified in vitro were confirmed in vivo using chemical profiling (SHALiPE-Seq); 2-methylnicotinic acid imidazolide (NAI) preferentially modifies the last Gs of G-tracts when RG4 is folded. They used a genetic approach to confirm the potential role of the RG4 structure in post-transcriptional gene regulation. The existence of RG4 was also confirmed in rice, suggesting the general presence of in vivo RG4s in the plant kingdom. (Summary by Mugdha Sabale) bioRxiv 10.1101/839621