Review: Plant networks as traits and hypotheses: Moving beyond description ($) (TIPS)
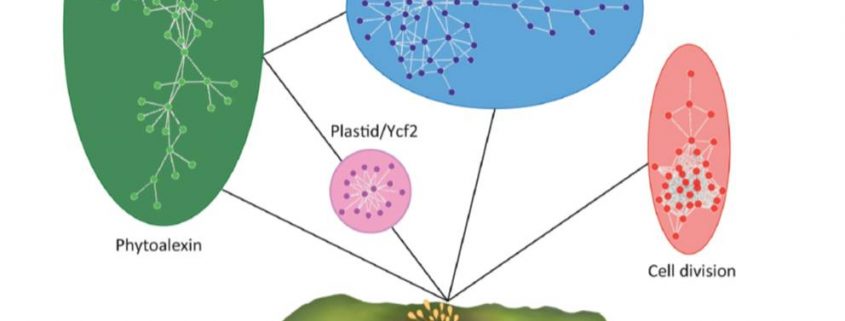
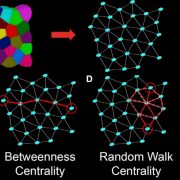
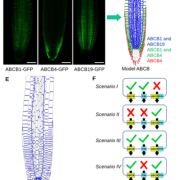
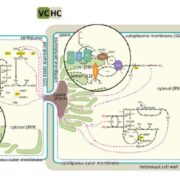
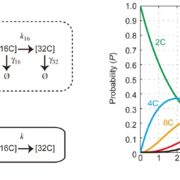
 In Star Wars Episode 2, Obi Wan identifies the location of a missing planet by walking through a 3D projection of the galaxy. I’ve always hoped that if we obtain enough data and figure out how to display it properly, we’ll “see” what parts are out of place or missing. But getting from simple linear models (“A leads to B, which leads to C”) to multidimensional models requires learning new ways of analyzing and presenting data. In this review, Marshall-Colón and Kliebenstein tackle this topic. They first introduce the idea that gene regulatory networks can be created from transcriptomic data, made even more illuminating when data from multiple time points are combined. They describe the usefulness of eigengenes, clusters of functionally-related genes that can be grouped for greater power. Of course, finding patterns within huge datasets requires the use of appropriate statistical tools as well as machine-learning approaches, several of which are presented here. Integrating across scales (e.g., genes and metabolites) lends additional power, as can integrating GWAS data with gene networks. The authors then describe how networks can produce testable hypotheses, using examples derived from nitrogen responses and specialized metabolite production. Finally, they observe the need for networks to be standardized for better sharing, and conclude by highlighting tools for enhanced visualization, including virtual reality. Perhaps Obi Wan’s world isn’t so far off. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.06.003
In Star Wars Episode 2, Obi Wan identifies the location of a missing planet by walking through a 3D projection of the galaxy. I’ve always hoped that if we obtain enough data and figure out how to display it properly, we’ll “see” what parts are out of place or missing. But getting from simple linear models (“A leads to B, which leads to C”) to multidimensional models requires learning new ways of analyzing and presenting data. In this review, Marshall-Colón and Kliebenstein tackle this topic. They first introduce the idea that gene regulatory networks can be created from transcriptomic data, made even more illuminating when data from multiple time points are combined. They describe the usefulness of eigengenes, clusters of functionally-related genes that can be grouped for greater power. Of course, finding patterns within huge datasets requires the use of appropriate statistical tools as well as machine-learning approaches, several of which are presented here. Integrating across scales (e.g., genes and metabolites) lends additional power, as can integrating GWAS data with gene networks. The authors then describe how networks can produce testable hypotheses, using examples derived from nitrogen responses and specialized metabolite production. Finally, they observe the need for networks to be standardized for better sharing, and conclude by highlighting tools for enhanced visualization, including virtual reality. Perhaps Obi Wan’s world isn’t so far off. (Summary by Mary Williams) Trends Plant Sci. 10.1016/j.tplants.2019.06.003