Review: Molecular concepts to explain heterosis in crops
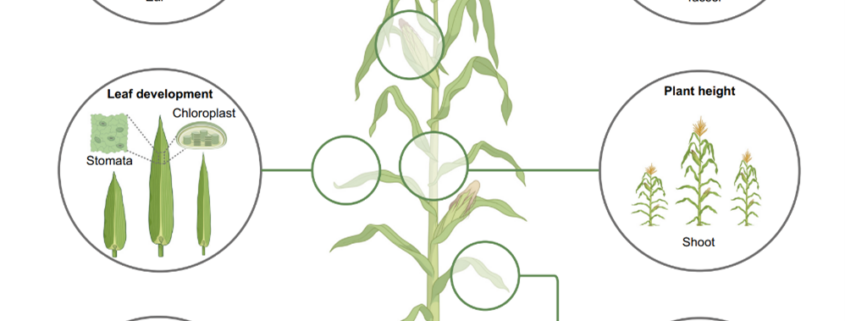
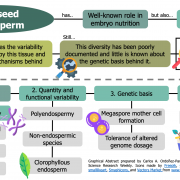
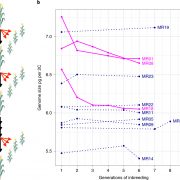
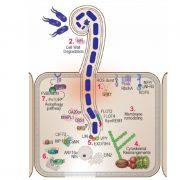
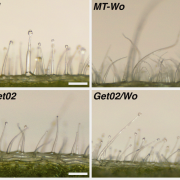
 Heterosis, the phenomenon where hybrid plants outperform their genetically distinct parents, is a cornerstone of modern agriculture. This review by Hochholdinger and Yu explores the molecular mechanisms behind heterosis. The review traces the history of heterosis, from its early discovery in tobacco by Kölreuter to its widespread use in crops like maize and rice. It covers classical genetic models of heterosis, including dominance, where superior dominant alleles mask recessive ones; overdominance, where heterozygous gene combinations outperform both homozygous forms; and epistasis, where interactions between multiple genes enhance hybrid traits, while integrating modern molecular insights into gene expression, epigenetic modifications, and protein regulation. The review discusses how significant genomic variation between parental lines drives genetic complementation in hybrid plants. This results in the activation of hundreds of additional genes, a process known as single-parent expression (SPE). Additionally, single-gene overdominance plays a significant role, where specific genes in hybrids enhance their vigor. The plant microbiome emerges as a critical factor in hybrid performance. Hybrids and their parental lines differ in microbial communities, particularly in the roots and leaves, influencing traits such as nutrient uptake and stress resistance. Beneficial soil microbes, acting as an extension of the plant genome, further enhance the growth and productivity of hybrids. One insight is the identification of metabolites as biomarkers for predicting heterosis. These metabolites, which fluctuate during critical growth stages, are indicators of traits like biomass and yield. Studies show that metabolite profiles can accurately predict hybrid performance across different environments, offering a powerful tool for breeders to optimize crop yields. (Summary by Amarachi Ezeoke) Trends Plant Sci 10.1016/j.tplants.2024.07.018
Heterosis, the phenomenon where hybrid plants outperform their genetically distinct parents, is a cornerstone of modern agriculture. This review by Hochholdinger and Yu explores the molecular mechanisms behind heterosis. The review traces the history of heterosis, from its early discovery in tobacco by Kölreuter to its widespread use in crops like maize and rice. It covers classical genetic models of heterosis, including dominance, where superior dominant alleles mask recessive ones; overdominance, where heterozygous gene combinations outperform both homozygous forms; and epistasis, where interactions between multiple genes enhance hybrid traits, while integrating modern molecular insights into gene expression, epigenetic modifications, and protein regulation. The review discusses how significant genomic variation between parental lines drives genetic complementation in hybrid plants. This results in the activation of hundreds of additional genes, a process known as single-parent expression (SPE). Additionally, single-gene overdominance plays a significant role, where specific genes in hybrids enhance their vigor. The plant microbiome emerges as a critical factor in hybrid performance. Hybrids and their parental lines differ in microbial communities, particularly in the roots and leaves, influencing traits such as nutrient uptake and stress resistance. Beneficial soil microbes, acting as an extension of the plant genome, further enhance the growth and productivity of hybrids. One insight is the identification of metabolites as biomarkers for predicting heterosis. These metabolites, which fluctuate during critical growth stages, are indicators of traits like biomass and yield. Studies show that metabolite profiles can accurately predict hybrid performance across different environments, offering a powerful tool for breeders to optimize crop yields. (Summary by Amarachi Ezeoke) Trends Plant Sci 10.1016/j.tplants.2024.07.018