Blocking the Guards: The ALY1 Nuclear Export Protein Is Required for DNA Methylation Machinery to Function
Plants constantly face the threat of attack from many directions. Organisms like bacteria, viruses, and fungi must be blocked from entering a plant’s cells, or quickly targeted for destruction once inside. Within the plant genome itself, transposable elements lie in wait for reactivation. In addition, a multitude of environmental conditions leave plants even more susceptible to these attacks. The genome isn’t defenseless, though, as plants have evolved several mechanisms to suppress invaders. The RNA-directed DNA Methylation (RdDM) pathway evolved as a broad means of plant defense. The RdDM pathway transcribes regions of repetitive DNA and generates small interfering RNAs (siRNAs) that can suppress transposable elements and transgenes, targeting these regions of DNA for methylation and guiding the formation of heterochromatin that can be epigenetically inherited.
In addition to RdDM, a second well-studied pathway important for cellular responses to stress involves the regulation of mRNA export from the nucleus to the cytoplasm (Katahira 2012). Choudury et al. (2019) have now connected the pathways for mRNA export and RdDM by identifying an mRNA export mutant necessary for RdDM. They focused on Arabidopsis thaliana ALY1, a member of a highly conserved gene family that participates in mRNA nuclear export in species ranging from yeast to humans. Since RdDM is largely responsible for cytosine methylation in the CHH context (where H = A, T, or G), the authors performed bisulfite-treated DNA sequencing on the aly1 mutant and discovered a global loss of methylation at RdDM CHH sites. The aly1 mutant had little effect on maintaining or copying existing methylation marks, affecting only methylation that is newly deposited during RdDM. This lack of RdDM resulted in the reactivation of some transposable elements and the inability of the mutant plants to de novo silence any new transgenes. Intriguingly, some siRNAs produced in the aly1 mutant are still able to leave the nucleus and enter the cytoplasm, indicating that the aly1 mutant must be blocking a different part of the RdDM pathway.
 To determine what is defective in the aly1 mutant, the authors used RNA immunoprecipitation sequencing (RIP-seq) to detect the interactions between ALY1 protein and any RNA bound to it. In line with ALY1 being a nuclear transporter, ALY1 bound several thousand unique mRNAs, including 325 high-confidence mRNAs. This result indicated that ALY1 binds a specific subset of mRNAs in Arabidopsis and that the ALY gene family (comprising four members in Arabidopsis: ALY1–ALY4) has partitioned its function across its several gene family members. None of the 325 high-confidence ALY1 RIP-seq targets included known RdDM components at first glance, though.
To determine what is defective in the aly1 mutant, the authors used RNA immunoprecipitation sequencing (RIP-seq) to detect the interactions between ALY1 protein and any RNA bound to it. In line with ALY1 being a nuclear transporter, ALY1 bound several thousand unique mRNAs, including 325 high-confidence mRNAs. This result indicated that ALY1 binds a specific subset of mRNAs in Arabidopsis and that the ALY gene family (comprising four members in Arabidopsis: ALY1–ALY4) has partitioned its function across its several gene family members. None of the 325 high-confidence ALY1 RIP-seq targets included known RdDM components at first glance, though.
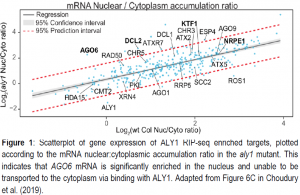
Given the expectation that the aly1 mutation leads to the nuclear accumulation of some mRNAs, the authors divided the mRNA from cells into nuclear and cytoplasmic fractions and performed mRNA-seq to detect which mRNAs are unable to be transported in aly1 mutants. Nearly 3,500 mRNAs showed mRNA export defects relative to wild type, significantly more than the 325 mRNAs identified as directly binding ALY1. By overlapping these genes with the RIP-seq targets, several mRNAs with known RdDM function were identified to export inefficiently in aly1 mutants. Most notably, ALY1 affected nuclear export of ARGONAUTE 6 (AGO6) mRNA (Figure 1), which should be translated into a protein that loads RdDM-derived small RNAs and carries them to their sites of action. AGO6 protein fails to accumulate in aly1 mutants, and complementation of the aly1 mutant with an overexpressing version of the AGO6 mRNA demonstrated that the signals for ALY1 function likely reside within the exons of the AGO6 mRNA.
Choudury et al. (2019) took a winding and elegant approach to an immensely complicated problem, weaving together the two well-tread fields of mRNA export and RdDM, eventually arriving at the AGO6 mRNA. This manuscript reiterates to us that linking genotype to phenotype benefits from a holistic molecular approach, with a deep consideration of gene family evolution, spatial gene expression, binding interactions, and a clever sense of how pathways are linked within a cell.
REFERENCES
Choudury, Sarah G., Saima Shahid, Diego Cuerda-Gil, Kaushik Panda, Alissa Cullen, Qurat Ul Ayn Ashraf, Meredith J. Sigman, Andrea D. McCue, and R. Keith Slotkin. 2019. “The RNA Export Factor ALY1 Enables Genome-Wide RNA-Directed DNA Methylation.” The Plant Cell. https://doi.org/10.1105/tpc.18.00624.
Katahira, Jun. 2012. “mRNA Export and the TREX Complex.” Biochimica et Biophysica Acta 1819 (6): 507–13.