Prediction of gene responses to heat and cold stress in maize
By Peng Zhou, University of Minnesota
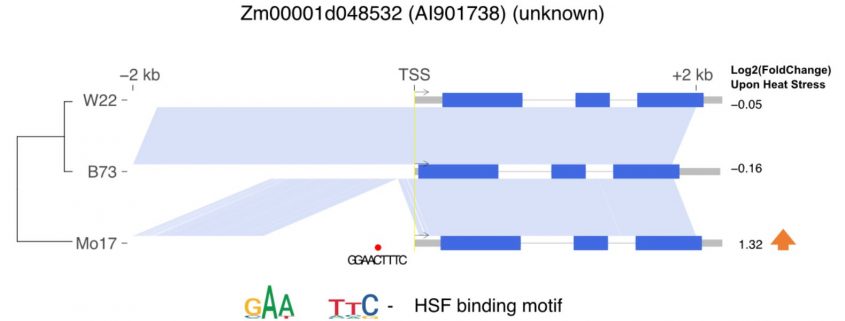
Background: Plants have developed sophisticated mechanisms to respond and acclimate to changing environmental conditions. Transcriptional regulation can occur in trans (by transcription factors, TFs) and/or in cis (transcription factor binding sites [TFBSs] in promoters). Within a population, studies have also revealed widespread variation both at the trans- (TF abundance) and cis- level (sequence difference at TFBSs). While it is straightforward to link TF abundance with their target gene responsiveness to stress, it remains largely unknown how sequence differences in cis-elements contribute to differences in a gene’s responsiveness to stress in general.
Question: Why do some genes respond to thermal stress in one genotype but not another? Is this “variable” response due in trans (abundance of its regulator TF) or in cis (promoter sequence difference)? Can we identify cis-regulatory motifs associated with stress-responsive expression patterns? Can these motifs be used to train a machine learning model to predict a gene responsiveness to stress?
Findings: Through three large-scale RNA-seq experiments, we systematically characterized the transcriptome response of maize seedlings to heat and cold stress. We identified many genes exhibiting altered expression patterns under stress between different maize genotypes, and assigned this expression variation to cis- or trans-regulatory mechanisms with the use of F1 hybrid data. We identified motifs associated with different stress-responsive patterns and used them to develop models to predict transcriptome responses. We highlight important parameters for motif detection and modeling of expression responses. We found both potential uses, and pitfalls, in how data from one genotype can be used to predict expression responses in other genotypes. Genes with variable response due to cis-regulatory variation highlight the importance of InDels and structural variants creating polymorphisms in key motifs in different haplotypes.
Next steps: The identification of known and novel cis-regulatory elements involved in stress response allows biotechnology applications such as design of stress-responsive promoters. There is also room for substantial improvement in model construction: by synthesizing more training data (larger genotype panel, more data types) and implementing more sophisticated training algorithm (convolutional neural networks), we hope to train models better at capturing the combinatorial logics of multiple cis-regulatory elements.
Reference:
Peng Zhou, Tara A. Enders, Zachary A. Myers, Erika Magnusson, Peter A Crisp, Jaclyn Noshay, Fabio Gomez-Cano, Zhikai Liang, Erich Grotewold, Kathleen Greenham, Nathan Springer (2022). Prediction of conserved and variable heat and cold stress response in maize using cis-regulatory information. The Plant Cell, https://doi.org/10.1093/plcell/koab267