Pol IV Function is Differentially Essential within the Brassicaceae
At least five RNA polymerases (Pol I to Pol V) are known to exist in plant cells. Pol IV produces short transcripts that are converted into double-stranded RNAs by RNA-dependent RNA polymerase 2 (RDR2) and processed into 24-nucleotide (nt) small interfering RNAs (siRNAs) by DICER-LIKE 3 (DCL3). These siRNAs direct ARGONAUTE 4 (AGO4) or AGO6 to sites of Pol V transcription and recruit DNA methyltransferases to initiate RNA-directed DNA methylation (RdDM) (reviewed in Cuerda-Gil and Slotkin, 2016). Information about Pol IV’s role in plant reproductive development is limited, in part due to the lack of visible mutant phenotypes in Arabidopsis (Arabidopsis thaliana), although it was recently demonstrated that it is critical for the triploid block in Arabidopsis (Satyaki and Gehring, 2019).
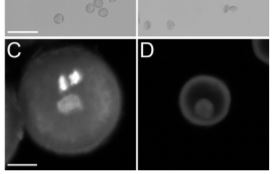
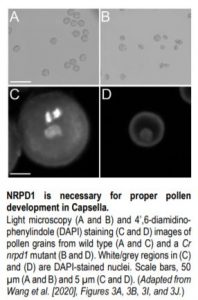
To examine the function of Pol IV in Capsella (Capsella rubella), a Brassicaceae species closely related to Arabidopsis, Wang et al. (2020) created a loss-of-function mutant of a major subunit of Capsella Pol IV, NRPD1, using CRISPR-Cas9. The mutant (named Cr nrpd1) exhibited obvious defects in female and male fertility. About 30% of the ovules of the homozygous Cr nrpd1 mutant remained unfertilized after pollination with wild-type pollen. When self-fertilized, the mutant produced seeds significantly reduced in size. These phenotypes resemble those of the recently reported nrpd1 mutant in Brassica rapa (Grover et al., 2019). In addition, pollen development in the homozygous Cr nrpd1 mutant was arrested at the post-meiotic, microspore stage (see figure).
 To understand the molecular basis of the pollen defect, the authors compared small RNA (sRNA) accumulation and transposable element (TE) expression profiles between wild-type and Cr nrpd1 microspores. The results showed that TE-derived 21- to 24-nt siRNAs are depleted in the Cr nrpd1 mutant, indicating that NRPD1, and thus Pol IV activity, is required for the biogenesis of these siRNAs. The extent of upregulation of TE expression positively correlates with the level of siRNAs associated with them, suggesting that the Pol IV-dependent siRNAs silence TEs in wild-type microspores. Moreover, these phenomena were also observed in an Arabidopsis nrpd1 mutant (At nrpd1), suggesting that the function of Pol IV in siRNA biogenesis is conserved between Capsella and Arabidopsis. Surprisingly, the authors also found in both species relatively high correlations (Spearman correlation coefficient = 0.69 and 0.77, respectively) between the abundances of TE-derived 24-nt and 21/22-nt siRNAs. This suggests that the different size classes of siRNAs are processed from shared precursors, and different DCL proteins compete for processing of the precursors to generate the different sizes of siRNAs. This hypothesis was supported by an analysis of publicly available sRNA sequencing data of an Arabidopsis mutant of DCL3 (the Dicer that produces 23/24-nt siRNAs) showing that the mutant exhibits elevated abundance of 21/22-nt siRNAs. Interestingly, RNA-seq analysis of Cr nrpd1 microspores detected enrichment of pollen tube growth-associated genes among the subsets of genes that showed downregulation in mRNA levels and associated 21/22-nt and 24-nt siRNA levels, suggesting a role of these genes in microspore/pollen development.
To understand the molecular basis of the pollen defect, the authors compared small RNA (sRNA) accumulation and transposable element (TE) expression profiles between wild-type and Cr nrpd1 microspores. The results showed that TE-derived 21- to 24-nt siRNAs are depleted in the Cr nrpd1 mutant, indicating that NRPD1, and thus Pol IV activity, is required for the biogenesis of these siRNAs. The extent of upregulation of TE expression positively correlates with the level of siRNAs associated with them, suggesting that the Pol IV-dependent siRNAs silence TEs in wild-type microspores. Moreover, these phenomena were also observed in an Arabidopsis nrpd1 mutant (At nrpd1), suggesting that the function of Pol IV in siRNA biogenesis is conserved between Capsella and Arabidopsis. Surprisingly, the authors also found in both species relatively high correlations (Spearman correlation coefficient = 0.69 and 0.77, respectively) between the abundances of TE-derived 24-nt and 21/22-nt siRNAs. This suggests that the different size classes of siRNAs are processed from shared precursors, and different DCL proteins compete for processing of the precursors to generate the different sizes of siRNAs. This hypothesis was supported by an analysis of publicly available sRNA sequencing data of an Arabidopsis mutant of DCL3 (the Dicer that produces 23/24-nt siRNAs) showing that the mutant exhibits elevated abundance of 21/22-nt siRNAs. Interestingly, RNA-seq analysis of Cr nrpd1 microspores detected enrichment of pollen tube growth-associated genes among the subsets of genes that showed downregulation in mRNA levels and associated 21/22-nt and 24-nt siRNA levels, suggesting a role of these genes in microspore/pollen development.
Nevertheless, why does the loss of Pol IV function cause defects in reproductive development in Capsella but not in Arabidopsis? The authors hypothesized that the different TE content in the two genomes (~24% and 50%, respectively) may explain their varied sensitivity to the disruption of Pol IV function. The authors found that the distances between TEs and neighboring genes are significantly shorter in Capsella than in Arabidopsis, suggesting that the reproductive defects in Capsella may be a consequence of TEs being located in close proximity to genes regulating/influencing pollen development, although future work will be necessary to further test this hypothesis.
This study greatly extends the findings from several other functional studies of Pol IV or RdDM-associated genes in rice, tomato, or Brassica rapa (reviewed in Chow et al., 2020), and provides novel insights into the roles of Pol IV in plant reproductive development that would be otherwise indiscernible in Arabidopsis, reinforcing the importance of investigating diverse plant species in future studies of Pol IV and RdDM.
Junpeng Zhan
Donald Danforth Plant Science Center
St. Louis, Missouri
JZhan@danforthcenter.org
ORCID ID: 0000-0001-7353-7608
REFERENCES
Chow, H.T., Chakraborty, T., and Mosher, R.A. (2020). RNA-directed DNA Methylation and sexual reproduction: expanding beyond the seed. Current opinion in plant biology 54: 11-17.
Cuerda-Gil, D., and Slotkin, R.K. (2016). Non-canonical RNA-directed DNA methylation. Nat Plants 2: 16163-16169.
Grover, J.W., Kendall, T., Baten, A., Burgess, D., Freeling, M., King, G.J., and Mosher, R.A. (2018). Maternal components of RNA-directed DNA methylation are required for seed development in Brassica rapa. Plant J 94: 575-582.
Satyaki, P.R.V., and Gehring, M. (2019). Paternally Acting Canonical RNA-Directed DNA Methylation Pathway Genes Sensitize Arabidopsis Endosperm to Paternal Genome Dosage. Plant Cell 31: 1563-1578.
Wang, Z., Butel, N., Santos-González, J., Borges, F., Yi, J., Martienssen, R.A., Martinez, G., Köhler, C. (2020). Polymerase IV Plays a Crucial Role in Pollen Development in Capsella. Plant Cell 32: 10.1105/tpc.19.00938.