Plant Science Research Weekly: November 1, 2024
Spotlight: The role of fossils for reconstructing the evolution of plant development
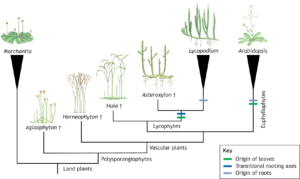 I suspect if we asked someone to describe a fossil we’d hear a lot about dinosaur bones. Certainly, science museums are full of fossilized animal remains, which have greatly informed our understanding of animal evolution. Plant fossils similarly are rich sources of information about plant evolution and evolutionary development (evo-devo), as highlighted here by Hetherington. The author starts with an observation that reconstructing evolution using only extant species misses out on all the stages that have become extinct, so can miss key forms and events (imagine trying to understand where birds came from without knowing about dinosaurs). He notes that piecing together plant evo-devo is challenging because most of the major innovations in plant form occurred 350 million years ago, well before the time when most terrestrial animals were around. He also makes the important point that if we only studied living plants, we would assume that leaves and roots each arise a single time, yet when we add in information from the fossil record it is apparent that the arose at least twice. This is a fascinating Spotlight article certainly worth sharing with students. (Summary by Mary Williams @PlantTeaching) Development 10.1242/dev.204322
I suspect if we asked someone to describe a fossil we’d hear a lot about dinosaur bones. Certainly, science museums are full of fossilized animal remains, which have greatly informed our understanding of animal evolution. Plant fossils similarly are rich sources of information about plant evolution and evolutionary development (evo-devo), as highlighted here by Hetherington. The author starts with an observation that reconstructing evolution using only extant species misses out on all the stages that have become extinct, so can miss key forms and events (imagine trying to understand where birds came from without knowing about dinosaurs). He notes that piecing together plant evo-devo is challenging because most of the major innovations in plant form occurred 350 million years ago, well before the time when most terrestrial animals were around. He also makes the important point that if we only studied living plants, we would assume that leaves and roots each arise a single time, yet when we add in information from the fossil record it is apparent that the arose at least twice. This is a fascinating Spotlight article certainly worth sharing with students. (Summary by Mary Williams @PlantTeaching) Development 10.1242/dev.204322
Perspective: Enzymatic routes to designer hemicelluloses for use in biobased materials
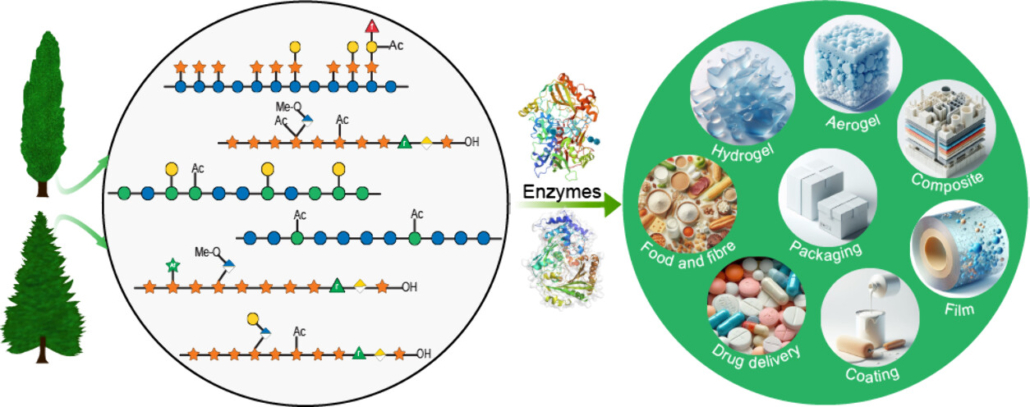 This article poses the interesting question of whether we can use our knowledge of plant cell wall-modifying, carbohydrate-active enzymes to produce biobased materials. Specifically, the authors point out that much of the hemicellulose contained in agricultural and wood fiber could provide a starting point for making useful products such as aerogels, films, and coatings. Compared to common chemical processes, enzymes can be both more specific about what is produced, and greener in terms of waste. This Perspective article gives a comprehensive review of the substrates and products of many different characterized enzymes (a good review for those interested) and also proposes steps to overcome some of the challenges inherent in creating designer hemicelluloses. For example, the authors propose screening enzymes for application rather than function, e.g., by measuring changes in sample viscosity or light-scattering, and they call for more collaboration between enzymologists and materials scientists. (Summary by Mary Williams @PlantTeaching) JACS Au 10.1021/jacsau.4c00469
This article poses the interesting question of whether we can use our knowledge of plant cell wall-modifying, carbohydrate-active enzymes to produce biobased materials. Specifically, the authors point out that much of the hemicellulose contained in agricultural and wood fiber could provide a starting point for making useful products such as aerogels, films, and coatings. Compared to common chemical processes, enzymes can be both more specific about what is produced, and greener in terms of waste. This Perspective article gives a comprehensive review of the substrates and products of many different characterized enzymes (a good review for those interested) and also proposes steps to overcome some of the challenges inherent in creating designer hemicelluloses. For example, the authors propose screening enzymes for application rather than function, e.g., by measuring changes in sample viscosity or light-scattering, and they call for more collaboration between enzymologists and materials scientists. (Summary by Mary Williams @PlantTeaching) JACS Au 10.1021/jacsau.4c00469
Review: Cracking the plant VOC sensing code and its practical applications
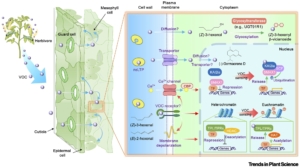 Many studies have demonstrated the importance of volatile organic compounds (VOCs) in communication between plants. VOCs emitted by a plant damaged by herbivory promote defenses in nearby plants. It is thought that these compounds may have originated as intra-plant signals, capable of moving long-distances faster than signals through the plant vascular system, with the inter-plant signaling function a fortuitous consequence. Notably, plants respond to signals produced by both conspecific and heterospecific individuals. A new review by Arimura and Uemura looks at these signals, reviewing their diverse functions but also highlighting what is known and not known about how they are perceived and elicit responses. Whether VOCs interact with specific plasma membrane-localized receptors (by analogy to animal olfactory system) remains an open question, and there is also some evidence that certain VOCs interact with proteins intracellularly. Some VOCs interact with the TOPLESS corepressor which may lead to changed in transcription and also chromatin remodeling. The review concludes with a discussion about how these insights might be used for protection of crop and horticultural plants, for example by the use of VOC-producing companion plants or synthetic VOCs. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.005
Many studies have demonstrated the importance of volatile organic compounds (VOCs) in communication between plants. VOCs emitted by a plant damaged by herbivory promote defenses in nearby plants. It is thought that these compounds may have originated as intra-plant signals, capable of moving long-distances faster than signals through the plant vascular system, with the inter-plant signaling function a fortuitous consequence. Notably, plants respond to signals produced by both conspecific and heterospecific individuals. A new review by Arimura and Uemura looks at these signals, reviewing their diverse functions but also highlighting what is known and not known about how they are perceived and elicit responses. Whether VOCs interact with specific plasma membrane-localized receptors (by analogy to animal olfactory system) remains an open question, and there is also some evidence that certain VOCs interact with proteins intracellularly. Some VOCs interact with the TOPLESS corepressor which may lead to changed in transcription and also chromatin remodeling. The review concludes with a discussion about how these insights might be used for protection of crop and horticultural plants, for example by the use of VOC-producing companion plants or synthetic VOCs. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.005
Review. Decoding resilience: Ecology, regulation, and evolution of biosynthetic gene clusters
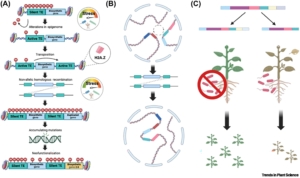 Although clusters of functionally related genes are common in prokaryotes, until recently it was thought that they were not a feature of eukaryotic genomes. However, several studies have identified biosynthetic gene clusters (BGCs) in plants. Many of these gene clusters include sets of enzymes that act sequentially in the production of specialized metabolites, such as defense or signaling molecules, enabling rapid and cost-effective production of the compound. A new review by Cawood and Ton discusses the function and regulation of these BGCs and speculates on how they are formed. Unlike prokaryotes, which have polycistronic BGCs (with many proteins encoded by a single mRNA), BGCs in eukaryotes are monocistronic, yet closely co-regulated, raising the question of how. Many studies suggest a role for histone modifications and histone variants (e.g., H2A.Z) in BGC co-regulation. There is also evidence for 3D chromatin topology and the formation of topologically associated domains (TADs) in BGC regulation. For example, in one case a chromatin loop structure leads to five BGC promoters simultaneously interacting with transcription factors and coactivators. Finally, the review turns to the question of how these BGCs were formed. It is generally assumed that they are derived from gene duplication and neofunctionalization, but the details of how this took place are not known. The authors describe a model in which stress induces activation of transposable elements, leading to gene duplication, exon shuffling, and gene fusion, providing opportunities for neofunctionalization. The authors also postulate that topologically associated domains could lead to related genes repositioning into BGCs. Finally, they note that in addition to positive selection of functional BGCs, there can be negative selection against incomplete BGCs due to the accumulation of potentially harmful metabolites. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.008
Although clusters of functionally related genes are common in prokaryotes, until recently it was thought that they were not a feature of eukaryotic genomes. However, several studies have identified biosynthetic gene clusters (BGCs) in plants. Many of these gene clusters include sets of enzymes that act sequentially in the production of specialized metabolites, such as defense or signaling molecules, enabling rapid and cost-effective production of the compound. A new review by Cawood and Ton discusses the function and regulation of these BGCs and speculates on how they are formed. Unlike prokaryotes, which have polycistronic BGCs (with many proteins encoded by a single mRNA), BGCs in eukaryotes are monocistronic, yet closely co-regulated, raising the question of how. Many studies suggest a role for histone modifications and histone variants (e.g., H2A.Z) in BGC co-regulation. There is also evidence for 3D chromatin topology and the formation of topologically associated domains (TADs) in BGC regulation. For example, in one case a chromatin loop structure leads to five BGC promoters simultaneously interacting with transcription factors and coactivators. Finally, the review turns to the question of how these BGCs were formed. It is generally assumed that they are derived from gene duplication and neofunctionalization, but the details of how this took place are not known. The authors describe a model in which stress induces activation of transposable elements, leading to gene duplication, exon shuffling, and gene fusion, providing opportunities for neofunctionalization. The authors also postulate that topologically associated domains could lead to related genes repositioning into BGCs. Finally, they note that in addition to positive selection of functional BGCs, there can be negative selection against incomplete BGCs due to the accumulation of potentially harmful metabolites. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci 10.1016/j.tplants.2024.09.008
PCMD: an interactive library for comparative metabolomics studies
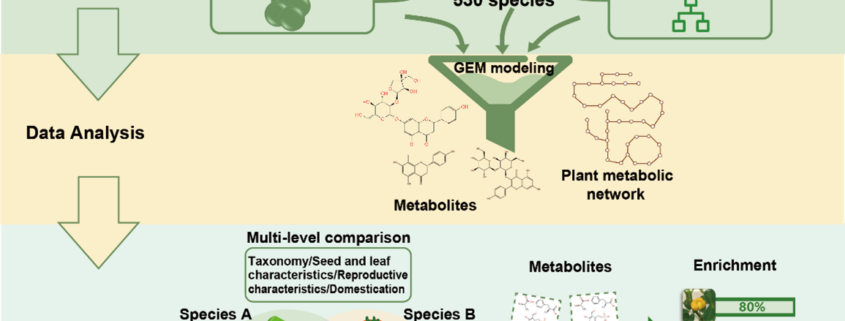
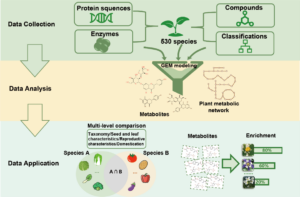 Albert Einstein once said, “The only thing that you absolutely have to know is the location of the library.” Libraries house vast troves of information for readers to explore, analyze, and use. With the exponential increase in data, libraries have also evolved into digital databases and online platforms. For example, large-scale omics studies yield tremendous amounts of data on genes, proteins, and metabolites that can help us understand plant development, provided they are accessible to researchers. In a recent issue of Plant Communications, Hu and colleagues introduced the Plant Comparative Metabolome Database (PCMD; https://yanglab.hzau.edu.cn/PCMD) for comparative metabolomics. Built on genome-based predictions of metabolites and associated metabolic pathways, along with supporting experimental data, PCMD provides metabolic profiles for 530 plant species. It offers unique features such as metabolite enrichment determination for cross-species comparative analysis, setting it apart from other databases. Each metabolite entry also includes data on associated proteins, metabolic reactions, and relevant literature, and links to databases like PubChem and MetaCyc. Future updates include tools for uploading experimental data and visualizing metabolic networks to deepen studies of gene-metabolite relationships. In conclusion, like a library, PCMD provides researchers interested in comparative metabolomics with a robust starting point for exploring metabolites, metabolic profiles, and the evolution of metabolic networks and can even support biotechnological research focused on plant-derived compounds for pharmaceutical or therapeutic applications. (Summary by Thomas Depaepe @thdpaepe) Plant Communications 10.1016/j.xplc.2024.101038.
Albert Einstein once said, “The only thing that you absolutely have to know is the location of the library.” Libraries house vast troves of information for readers to explore, analyze, and use. With the exponential increase in data, libraries have also evolved into digital databases and online platforms. For example, large-scale omics studies yield tremendous amounts of data on genes, proteins, and metabolites that can help us understand plant development, provided they are accessible to researchers. In a recent issue of Plant Communications, Hu and colleagues introduced the Plant Comparative Metabolome Database (PCMD; https://yanglab.hzau.edu.cn/PCMD) for comparative metabolomics. Built on genome-based predictions of metabolites and associated metabolic pathways, along with supporting experimental data, PCMD provides metabolic profiles for 530 plant species. It offers unique features such as metabolite enrichment determination for cross-species comparative analysis, setting it apart from other databases. Each metabolite entry also includes data on associated proteins, metabolic reactions, and relevant literature, and links to databases like PubChem and MetaCyc. Future updates include tools for uploading experimental data and visualizing metabolic networks to deepen studies of gene-metabolite relationships. In conclusion, like a library, PCMD provides researchers interested in comparative metabolomics with a robust starting point for exploring metabolites, metabolic profiles, and the evolution of metabolic networks and can even support biotechnological research focused on plant-derived compounds for pharmaceutical or therapeutic applications. (Summary by Thomas Depaepe @thdpaepe) Plant Communications 10.1016/j.xplc.2024.101038.
The ”hourglass” model of embryogenesis extends to brown algae
 The hourglass model of embryogenesis was proposed in the 1990s, and extended to green plants and fungi in the 2010s. During animal embryogenesis, the very earliest stages (post fertilization) are morphologically quite different from each other, and the later stages are quite different, but in the middle of embryogenesis there is a period, called the phylotypic period, at which the appearance of animal embryos converge and look quite similar. This pattern of morphological diversity resembles the shape of an hourglass, broad at the top and bottom and narrow in the middle. The phylotypic period is the period during which the basic body plan of the organism is established. Interestingly, during the phylotypic period, there is also a narrowing of patterns of gene expression, and a shift toward the expression of evolutionarily older, more highly conserved genes. In a new paper, Lotharukpong et al. found evidence for this hourglass model of development in another type of multicellular organism, the brown algae, which became multicellular independently of plants, fungi and animals. The study found that during the phylotypic period when the algal body plan was being established, younger genes were expressed less, rather than older genes being expressed more, and the more ancient genes expressed during this stage are also more pleiotropically expressed than younger genes, consistent with findings from animals and plants. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-024-08059-8
The hourglass model of embryogenesis was proposed in the 1990s, and extended to green plants and fungi in the 2010s. During animal embryogenesis, the very earliest stages (post fertilization) are morphologically quite different from each other, and the later stages are quite different, but in the middle of embryogenesis there is a period, called the phylotypic period, at which the appearance of animal embryos converge and look quite similar. This pattern of morphological diversity resembles the shape of an hourglass, broad at the top and bottom and narrow in the middle. The phylotypic period is the period during which the basic body plan of the organism is established. Interestingly, during the phylotypic period, there is also a narrowing of patterns of gene expression, and a shift toward the expression of evolutionarily older, more highly conserved genes. In a new paper, Lotharukpong et al. found evidence for this hourglass model of development in another type of multicellular organism, the brown algae, which became multicellular independently of plants, fungi and animals. The study found that during the phylotypic period when the algal body plan was being established, younger genes were expressed less, rather than older genes being expressed more, and the more ancient genes expressed during this stage are also more pleiotropically expressed than younger genes, consistent with findings from animals and plants. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-024-08059-8
Single-plant omics provides transcriptional insights into the transition from the vegetative to reproductive phases
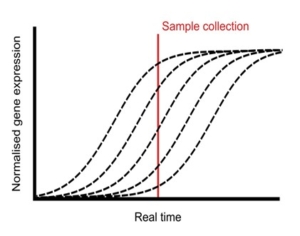 Plants undergo a series of physiological processes when transitioning from the juvenile to the vegetative phase, and then vegetative to the reproductive phase. RNA-Seq offers substantial potential for uncovering the transcriptional landscape underlying these developmental transitions. However, developmental asynchrony among individual plants within a population creates variations in the spatiotemporal expression of genes. Redmond et al. performed a single-plant-omics study on a large population of Arabidopsis thaliana and their investigation revealed the detailed sequence of transcriptional events that occur before and after the bolting transition. Using single-plant-omics allowed the researchers to order individual plants by their intrinsic biological age, providing a high-resolution transcriptional landscape. They trained different models to test whether these models are more or less informative compared to gene expression data. Results revealed that most of the differentially expressed genes were closely linked with the traits of biomass and leaf area. Implication of the pseudotime inference algorithm revealed certain senescence-related traits e.g., transcriptional repression of ribosome biosynthesis followed by photosynthesis shutdown is a major event that serves as a landmark when a bolting plant changes from vegetative to reproductive phase. Their study highlights the role of environmental asynchrony and opens new avenues to use single-plant-omics in synthetic biology and molecular breeding programs. (Summary by Asif Ali @pbgasifkalas) Plant Cell. 10.1093/plcell/koae226
Plants undergo a series of physiological processes when transitioning from the juvenile to the vegetative phase, and then vegetative to the reproductive phase. RNA-Seq offers substantial potential for uncovering the transcriptional landscape underlying these developmental transitions. However, developmental asynchrony among individual plants within a population creates variations in the spatiotemporal expression of genes. Redmond et al. performed a single-plant-omics study on a large population of Arabidopsis thaliana and their investigation revealed the detailed sequence of transcriptional events that occur before and after the bolting transition. Using single-plant-omics allowed the researchers to order individual plants by their intrinsic biological age, providing a high-resolution transcriptional landscape. They trained different models to test whether these models are more or less informative compared to gene expression data. Results revealed that most of the differentially expressed genes were closely linked with the traits of biomass and leaf area. Implication of the pseudotime inference algorithm revealed certain senescence-related traits e.g., transcriptional repression of ribosome biosynthesis followed by photosynthesis shutdown is a major event that serves as a landmark when a bolting plant changes from vegetative to reproductive phase. Their study highlights the role of environmental asynchrony and opens new avenues to use single-plant-omics in synthetic biology and molecular breeding programs. (Summary by Asif Ali @pbgasifkalas) Plant Cell. 10.1093/plcell/koae226
Phloem loading and subcellular transport drive carbon storage in cassava roots
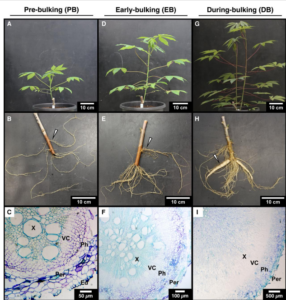 Cassava (Manihot esculenta) is a vital starchy crop essential for food security in Sub-Saharan Africa, South America, and Southeast Asia. A recent study on cassava by Rüscher et al. provides important insights into the plant’s sugar control mechanisms as the roots expand, produce large amounts of storage parenchyma, and accumulate sugars and starch, a process known as root bulking. The authors examined how carbohydrates are transported into the roots and found that both the absence of upregulation of specific transporters and the presence of branched plasmodesmata support a model of passive symplastic phloem loading. They also found evidence of subcellular compartmentalization of sugars in the storage root during bulking. As sugar levels build in the cytosol, monosaccharide transporter genes that import sugars into the vacuole were more highly expressed, leading to vacuoles in storage roots containing up to 7.5% sucrose by dry weight. The accumulation of sugars establishes an osmotic gradient that attracts water into the vacuoles, resulting in the enlargement of root cells and potentially affecting the plant’s ability to regulate internal water resources. By understanding the dynamics of sugar compartmentation, gene expression, and transport patterns, the study highlights an immense opportunity for cassava breeding projects. (Summary by Tuyelee Das @das_tuyelee) Plant Physiol. 10.1093/plphys/kiae298
Cassava (Manihot esculenta) is a vital starchy crop essential for food security in Sub-Saharan Africa, South America, and Southeast Asia. A recent study on cassava by Rüscher et al. provides important insights into the plant’s sugar control mechanisms as the roots expand, produce large amounts of storage parenchyma, and accumulate sugars and starch, a process known as root bulking. The authors examined how carbohydrates are transported into the roots and found that both the absence of upregulation of specific transporters and the presence of branched plasmodesmata support a model of passive symplastic phloem loading. They also found evidence of subcellular compartmentalization of sugars in the storage root during bulking. As sugar levels build in the cytosol, monosaccharide transporter genes that import sugars into the vacuole were more highly expressed, leading to vacuoles in storage roots containing up to 7.5% sucrose by dry weight. The accumulation of sugars establishes an osmotic gradient that attracts water into the vacuoles, resulting in the enlargement of root cells and potentially affecting the plant’s ability to regulate internal water resources. By understanding the dynamics of sugar compartmentation, gene expression, and transport patterns, the study highlights an immense opportunity for cassava breeding projects. (Summary by Tuyelee Das @das_tuyelee) Plant Physiol. 10.1093/plphys/kiae298