Plant Science Research Weekly: March 8th
Review: Evolutionary and ecological insights from herbicide resistant weeds: what have we learned about plant adaptation, and what is left to uncover?
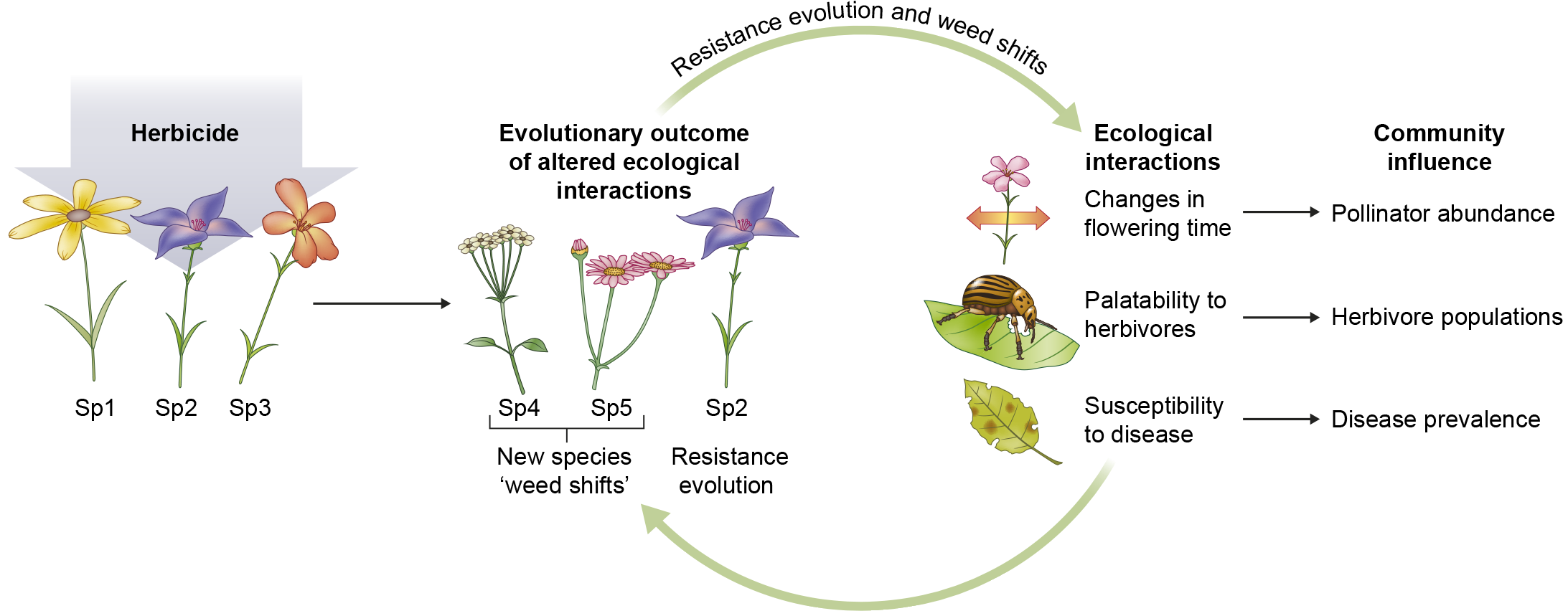 Weeds represent a problem for the economy due to their easy resistance of pesticides, but they are also a model for adaptation, ecology and evolution. In this review Baucom focusses on three areas: the genetic basis of adaptation, evolutionary constraints, and experimental evolution. Weed resistance is related to mutations of target site resistance genes (e.g., ALS gene, encoding acetolactate synthase, conferring resistance to ALS-inhibiting herbicides), and genes involved in herbicide detoxification (e.g., glutathione S-transferases). Resistance phenotypes are often shared in other lineages with the same genetic changes due to convergent evolution (e.g., duplications in EPSPS gene in four different species leading to glyphosate resistance). Resistance phenotypes can be related to optimal pleiotropy, where some mutations of genes are more common than others because of their effect on fitness. The author discusses experimental evolution as a tool to know if weed populations can evolve higher levels of resistance; low doses of herbicides applied in Lolium rigidum has the consequence that in two or four generations plants were resistant. The author also addresses how herbicide resistance affects ecological interactions, for example with pollinators. The author summarizes “Here I have attempted to emphasize that the plant/herbicide model is uniquely poised to address questions in evolution and ecology more broadly”. (Summary by Laura Alejandra Mejía Agudelo) New Phytol. 10.1111/nph.15723
Weeds represent a problem for the economy due to their easy resistance of pesticides, but they are also a model for adaptation, ecology and evolution. In this review Baucom focusses on three areas: the genetic basis of adaptation, evolutionary constraints, and experimental evolution. Weed resistance is related to mutations of target site resistance genes (e.g., ALS gene, encoding acetolactate synthase, conferring resistance to ALS-inhibiting herbicides), and genes involved in herbicide detoxification (e.g., glutathione S-transferases). Resistance phenotypes are often shared in other lineages with the same genetic changes due to convergent evolution (e.g., duplications in EPSPS gene in four different species leading to glyphosate resistance). Resistance phenotypes can be related to optimal pleiotropy, where some mutations of genes are more common than others because of their effect on fitness. The author discusses experimental evolution as a tool to know if weed populations can evolve higher levels of resistance; low doses of herbicides applied in Lolium rigidum has the consequence that in two or four generations plants were resistant. The author also addresses how herbicide resistance affects ecological interactions, for example with pollinators. The author summarizes “Here I have attempted to emphasize that the plant/herbicide model is uniquely poised to address questions in evolution and ecology more broadly”. (Summary by Laura Alejandra Mejía Agudelo) New Phytol. 10.1111/nph.15723
Review: Alternate grassy ecosystem states are determined by palatability-flammability trade-offs ($)
 Grassy ecosystems tend towards two stable states. One, the grazing-lawn state, is dominated by shoter grasses that thrive when grazed by herbivores, and the other, the fire-grass state, is dominated by taller grasses thrive when periodically controlled by burning. The flux between fire-grass and grazing-lawn ecosystems can be attributed to the positive feedbacks promoted by the respective environmentally-adaptive traits of the inhabitant grasses. Since grazing-lawn grasses grow low to the ground, grazing is necessary to provide adequate light. These grasses promote grazing by offering ample nutritious and easy-to-forage leaf material, which in turn allows stems and roots to remain unharmed and continue to proliferate. Fire-grasses are sturdy and resist decomposition, resulting in a buildup of dead biomass that shades competitors and provides fuel for fires. Fire-grasses are well protected from fires and are quick to reestablish and proliferate after a burn, unlike their competitors. Although the underlying systems are different, the dynamics of the fire-grass vs. grazing-lawn and forest vs. savanna alternate stable states both display the impact initial conditions and lag effects have on ecological succession. A precise description of the cycle between these ecosystem states is yet unknown, however the importance of grasses and their varied traits is apparent. (Summary by Rebecca Hayes) Trends Ecol. Evol. 10.1016/j.tree.2019.01.007
Grassy ecosystems tend towards two stable states. One, the grazing-lawn state, is dominated by shoter grasses that thrive when grazed by herbivores, and the other, the fire-grass state, is dominated by taller grasses thrive when periodically controlled by burning. The flux between fire-grass and grazing-lawn ecosystems can be attributed to the positive feedbacks promoted by the respective environmentally-adaptive traits of the inhabitant grasses. Since grazing-lawn grasses grow low to the ground, grazing is necessary to provide adequate light. These grasses promote grazing by offering ample nutritious and easy-to-forage leaf material, which in turn allows stems and roots to remain unharmed and continue to proliferate. Fire-grasses are sturdy and resist decomposition, resulting in a buildup of dead biomass that shades competitors and provides fuel for fires. Fire-grasses are well protected from fires and are quick to reestablish and proliferate after a burn, unlike their competitors. Although the underlying systems are different, the dynamics of the fire-grass vs. grazing-lawn and forest vs. savanna alternate stable states both display the impact initial conditions and lag effects have on ecological succession. A precise description of the cycle between these ecosystem states is yet unknown, however the importance of grasses and their varied traits is apparent. (Summary by Rebecca Hayes) Trends Ecol. Evol. 10.1016/j.tree.2019.01.007
Evolution of chloroplast retrograde signaling facilitates green plant adaptation to land
 The evolution of signaling pathways in plants enabled the water to land transition, during which drought response was crucial for the adaptation to terrestrial habitats. Here, Zhao et al. reveal the role of drought- induced phosphoadenosine (PAP) in the regulation of abscisic acid (ABA) synthesis in the chloroplasts, which leads to stomatal closure. Also, they found PAP is mediated by the nucleotide phosphatase SAL1. The authors postulate “PAP-induced stomatal closure, guard-cell ROS and NO signaling, and ion transport” are evolutionarily conserved, by using stomatal bioassays, fluorescent indicators, bioinformatics and comparative genetic similarity analysis comparing 31 species including land plants, streptophyte and chlorophyte algae. Furthermore, this study suggests SAL-PAP retrograde signaling pathway and ABA signaling evolved in streptophytes prior to stomata formation in the process of terrestrialization. This work provides insights into the evolution of signaling pathways which leads to crops capable to adapt to water scarcity conditions. (Summary by Ana Valladares) PNAS 10.1073/pnas.1812092116
The evolution of signaling pathways in plants enabled the water to land transition, during which drought response was crucial for the adaptation to terrestrial habitats. Here, Zhao et al. reveal the role of drought- induced phosphoadenosine (PAP) in the regulation of abscisic acid (ABA) synthesis in the chloroplasts, which leads to stomatal closure. Also, they found PAP is mediated by the nucleotide phosphatase SAL1. The authors postulate “PAP-induced stomatal closure, guard-cell ROS and NO signaling, and ion transport” are evolutionarily conserved, by using stomatal bioassays, fluorescent indicators, bioinformatics and comparative genetic similarity analysis comparing 31 species including land plants, streptophyte and chlorophyte algae. Furthermore, this study suggests SAL-PAP retrograde signaling pathway and ABA signaling evolved in streptophytes prior to stomata formation in the process of terrestrialization. This work provides insights into the evolution of signaling pathways which leads to crops capable to adapt to water scarcity conditions. (Summary by Ana Valladares) PNAS 10.1073/pnas.1812092116
Synergistic pectin degradation and guard cell pressurization underlie stomatal pore formation

This is a very interesting paper published by Rui et al. on the molecular mechanisms for stomatal pore formation, which includes pore initiation followed by pore enlargement. Each pore is surrounded by a pair of guard cells, which regulate the aperture of the pore and control gas exchange and water loss between the plant and the environment. The cellular details of the local separation between sister guard cells that give rise to the stomatal pore, or how formation of this pore is reached, are not yet fully understood. Stomatal pore initiation is an extremely dynamic process in Arabidopsis, with several morphological traits observed in a 3D time lapse imaging. The authors describe that an increase of pectic homogalacturonan (HG) modification promotes stomatal pore formation, and the inhibition of the HG de-methyl-esterification delays pore initiation and suppress pore enlargement. Moreover, there is evidence for a role of turgor pressure in stomatal pore formation. Specifically, cytokinesis could be prevented with hyperosmotic treatment in guard mother cells, and in cells treated after cytokinesis a reduction of pore enlargement was observed. As the authors conclude, “These data provide evidence that HG delivery and modification, and guard cell pressurization, make functional contributions to stomatal pore initiation and enlargement.” (Summarized by Francesca Resentini) Plant Physiol. 10.1104/pp.19.00135
An aberrant protein phosphatase 2C confers abscisic acid tolerance and drives high transpiration during drought conditions in a parasitic plant, Striga ($)
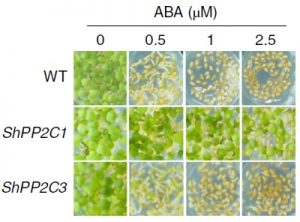 Striga hermonthica is a parasitic plant which infects major crops in arid environments. The rate of transpiration in Striga is higher than that of the host plant, thus maintaining a water potential gradient from the host to the Striga plant. Until recently, the exact mechanism has been poorly understood. A recent study from Fujioka et al, uncovered the mechanism which underlies this phenomenon, namely that Striga plants do not respond to abscisic acid (ABA), the hormone responsible for stomatal closure, resulting in high transpiration even in low water conditions. Furthermore, the authors identified that a Striga protein phosphatase 2C (ShPP2C1) possesses mutations which inhibits its ability to interact with ABA. ShPP2C1 was transformed into Arabidopsis resulting in plants which were insensitive to ABA, indicating that the ShPP2C1 acts dominantly to negatively regulate ABA signaling. Taken together, these results present the mechanism behind Striga infections and offer insights into possible targets for preventing major crop losses caused by Striga. (Summary by Nick Segerson) Nature Plants 10.1038/s41477-019-0362-7
Striga hermonthica is a parasitic plant which infects major crops in arid environments. The rate of transpiration in Striga is higher than that of the host plant, thus maintaining a water potential gradient from the host to the Striga plant. Until recently, the exact mechanism has been poorly understood. A recent study from Fujioka et al, uncovered the mechanism which underlies this phenomenon, namely that Striga plants do not respond to abscisic acid (ABA), the hormone responsible for stomatal closure, resulting in high transpiration even in low water conditions. Furthermore, the authors identified that a Striga protein phosphatase 2C (ShPP2C1) possesses mutations which inhibits its ability to interact with ABA. ShPP2C1 was transformed into Arabidopsis resulting in plants which were insensitive to ABA, indicating that the ShPP2C1 acts dominantly to negatively regulate ABA signaling. Taken together, these results present the mechanism behind Striga infections and offer insights into possible targets for preventing major crop losses caused by Striga. (Summary by Nick Segerson) Nature Plants 10.1038/s41477-019-0362-7
NERD1, a novel regulator of ovule number in Arabidopsis
 Seed number is a critical component of crop yield; the number of ovules determines the number of seeds. Ovule initiation in the carpel margin meristem (CMM) is controlled by genetic (i.e., AINTEGUMENTA, LEUNIG, SEUSS) and hormonal factors (auxins, cytokinins, gibberellins, and brassinosteroids). Most of the mutants affected in ovule number have also defects in other tissues. In this study, Yuan and Kessler wanted to identify actors that could regulate ovule number without affecting other tissues. The authors exploited natural variation in ovule number in 189 Arabidopsis accessions. They conducted a Genome Wide Association Study (GWAS) and identified some genomic regions linked to variation in ovule number. Then they focused in the genes containing the most significantly associated SNPs and analyzed mutants for those genes. Insertional lines for the NEW ENHANCER of ROOT DWARFISM (NERD1) gene showed reduced ovule number, ovule defects, and other male defects; while overexpression of NERD1 in the Altair-5 background (a low number accession) increased ovule number. Interestingly, the NERD1 gene is expressed ubiquitously during Arabidopsis development; the NERD1 is predicted to be a membrane protein. Further work is required to establish at which level NERD1 interacts with the pathways controlling ovule initiation. This research extends our knowledge of how ovule number (and hence seed number) could be increased. (Summary by Humberto Herrera-Ubaldo) PLOS Genetics 10.1371/journal.pgen.1007934
Seed number is a critical component of crop yield; the number of ovules determines the number of seeds. Ovule initiation in the carpel margin meristem (CMM) is controlled by genetic (i.e., AINTEGUMENTA, LEUNIG, SEUSS) and hormonal factors (auxins, cytokinins, gibberellins, and brassinosteroids). Most of the mutants affected in ovule number have also defects in other tissues. In this study, Yuan and Kessler wanted to identify actors that could regulate ovule number without affecting other tissues. The authors exploited natural variation in ovule number in 189 Arabidopsis accessions. They conducted a Genome Wide Association Study (GWAS) and identified some genomic regions linked to variation in ovule number. Then they focused in the genes containing the most significantly associated SNPs and analyzed mutants for those genes. Insertional lines for the NEW ENHANCER of ROOT DWARFISM (NERD1) gene showed reduced ovule number, ovule defects, and other male defects; while overexpression of NERD1 in the Altair-5 background (a low number accession) increased ovule number. Interestingly, the NERD1 gene is expressed ubiquitously during Arabidopsis development; the NERD1 is predicted to be a membrane protein. Further work is required to establish at which level NERD1 interacts with the pathways controlling ovule initiation. This research extends our knowledge of how ovule number (and hence seed number) could be increased. (Summary by Humberto Herrera-Ubaldo) PLOS Genetics 10.1371/journal.pgen.1007934
Cell type specific transcriptional reprogramming during Ustilago maydis and maize interaction
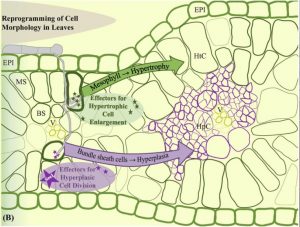
Figure from Redkar et al. 2017
Ustilago maydis is a model biotrophic fungus which causes smut disesase in maize, characterized by tumorous symptoms on all aerial parts. Tumor formation in leaves of the host is a result of massive reprogramming by modulation of two processes, hypertrophy (cell expansion) and hyperplasia (cell division). Previously, it has been shown that the tumor formation is mediated in a cell-type specific manner. Mesophyll cells show hypertrophy, while hyperplasia occurs exclusively in the bundle sheath cells. Recently, Villajuana-Bonequi et al. characterized the plant transcriptional program that occurs in tumorous mesophyll and bundle sheath cells using a combination of techniques such as high resolution laser capture microdissection and cell specific RNAseq. In this study they observed that genes involved in cell cycle regulation get altered in tumorous cells. Hence, the study provides interesting insights to understand tumor formation by plant pathogens at cellular resolution. (Summary by Mugdha Sabale) bioRxiv 10.1101/552562v1
Two opposing pathways independently regulate symbiotic nodulation in Medicago truncatula ($)
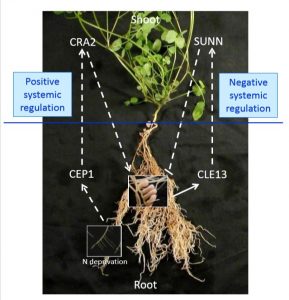 To optimize usage of nutrient, legumes utilize multiple strategies to regulate root nitrogen-fixing symbiotic nodulation. Excessive nodulation may hamper plant’s normal growth, so fine-tuning it is important for plant survival. Laffont et al. report that there are two independent pathways systematically regulating symbiotic nodulation in Medicago truncatula. They found that Super Numeric Nodule (SUNN) and Compact Root Architecture 2 (CRA2) systemic pathways act antagonistically and lead to weakened and enhanced nodulation capacity of roots, respectively. However, they also showed that these two pathways have little genetic interaction and act independently. This study shows that Medicago truncatula has evolved a sophisticated mechanism to balance nutrient distribution in the symbiotic relationship between itself and rhizobia. (Summarized by Nanxun Qin) Plant Physiology 10.1104/pp.18.01588
To optimize usage of nutrient, legumes utilize multiple strategies to regulate root nitrogen-fixing symbiotic nodulation. Excessive nodulation may hamper plant’s normal growth, so fine-tuning it is important for plant survival. Laffont et al. report that there are two independent pathways systematically regulating symbiotic nodulation in Medicago truncatula. They found that Super Numeric Nodule (SUNN) and Compact Root Architecture 2 (CRA2) systemic pathways act antagonistically and lead to weakened and enhanced nodulation capacity of roots, respectively. However, they also showed that these two pathways have little genetic interaction and act independently. This study shows that Medicago truncatula has evolved a sophisticated mechanism to balance nutrient distribution in the symbiotic relationship between itself and rhizobia. (Summarized by Nanxun Qin) Plant Physiology 10.1104/pp.18.01588
Crop variety management for climate adaptation supported by citizen science
 Current research is focused on developing better crop varieties that could withstand the adversities posed by climate change; however, these varieties might perform worse than the locally-grown varieties in field conditions. This could be due to the release of varieties to farmers without performing geographic analysis and without considering the local climatic as well as production conditions. To address this, van Etten et al. have involved the farmers as ‘citizen scientists’ to evaluate the relative performance of new varieties that were randomly assigned to them. The authors have organized tricot (triadic comparisons of technologies) trials covering 842 plots of common bean in Nicaragua (five cropping seasons), 1090 plots of durum wheat in Ethiopia (three seasons), and 10477 plots of bread wheat in India (four seasons) between 2012 and 2016. Modeling the environmental influence (agroclimatic and soil data) on variety performance has pinpointed the varieties that are better adapted to local climates. The study has highlighted the possibilities to provide geographically specific recommendations to improve crop adaptation and reduce the risk of farmers facing the adverse effects of climate change. (Summary by Muthamilarasan Mehanathan) PNAS 10.1073/pnas.1813720116
Current research is focused on developing better crop varieties that could withstand the adversities posed by climate change; however, these varieties might perform worse than the locally-grown varieties in field conditions. This could be due to the release of varieties to farmers without performing geographic analysis and without considering the local climatic as well as production conditions. To address this, van Etten et al. have involved the farmers as ‘citizen scientists’ to evaluate the relative performance of new varieties that were randomly assigned to them. The authors have organized tricot (triadic comparisons of technologies) trials covering 842 plots of common bean in Nicaragua (five cropping seasons), 1090 plots of durum wheat in Ethiopia (three seasons), and 10477 plots of bread wheat in India (four seasons) between 2012 and 2016. Modeling the environmental influence (agroclimatic and soil data) on variety performance has pinpointed the varieties that are better adapted to local climates. The study has highlighted the possibilities to provide geographically specific recommendations to improve crop adaptation and reduce the risk of farmers facing the adverse effects of climate change. (Summary by Muthamilarasan Mehanathan) PNAS 10.1073/pnas.1813720116



