Plant Science Research Weekly: March 7, 2025
Review: The complexities of metabolite transport in C4 photosynthesis
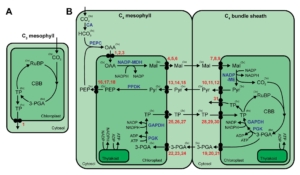 C4 photosynthesis, an adaptive mechanism to spatially concentrate CO2 around Rubisco to enhance carbon fixation, has evolved independently at least 60 times in plants. This process spatially separates the initial carbon fixation by PEPC and carbon reduction by Rubisco, which requires that compounds move in and out of various organelles of mesophyll cells and bundle sheath cells. While scientists have made great strides in bridging the gap in our understanding of photosynthesis evolution, there are still questions that need to be answered in terms of metabolite transport. This comprehensive review by Mattison and Kelly presents our current understanding of these important transporters, highlighting that much of what is known about them is derived from studies in C3 plants. They point to several critical unknowns; for example, the transporter facilitating H+-dependent pyruvate uptake in several species including maize is not yet known. Similarly, the transporters that move malate and pyruvate in the bundle sheath chloroplast of NADP-ME subtype C4 species are also unknown. It is also hypothesized that an unidentified aspartate transporter is involved in malate transport to the chloroplast. There is a great need for more experimental validation of metabolite transporters in C4 plants. With continuous advances in molecular, synthetic, genomic biology and genetic engineering, researchers are working to elucidate the complete metabolite transport mechanisms in C4 crops, which is necessary in order to improve photosynthesis and water-use efficiency in C3 crops. (Summary by Mae Mercado) The Plant Cell 10.1093/plcell/koaf019
C4 photosynthesis, an adaptive mechanism to spatially concentrate CO2 around Rubisco to enhance carbon fixation, has evolved independently at least 60 times in plants. This process spatially separates the initial carbon fixation by PEPC and carbon reduction by Rubisco, which requires that compounds move in and out of various organelles of mesophyll cells and bundle sheath cells. While scientists have made great strides in bridging the gap in our understanding of photosynthesis evolution, there are still questions that need to be answered in terms of metabolite transport. This comprehensive review by Mattison and Kelly presents our current understanding of these important transporters, highlighting that much of what is known about them is derived from studies in C3 plants. They point to several critical unknowns; for example, the transporter facilitating H+-dependent pyruvate uptake in several species including maize is not yet known. Similarly, the transporters that move malate and pyruvate in the bundle sheath chloroplast of NADP-ME subtype C4 species are also unknown. It is also hypothesized that an unidentified aspartate transporter is involved in malate transport to the chloroplast. There is a great need for more experimental validation of metabolite transporters in C4 plants. With continuous advances in molecular, synthetic, genomic biology and genetic engineering, researchers are working to elucidate the complete metabolite transport mechanisms in C4 crops, which is necessary in order to improve photosynthesis and water-use efficiency in C3 crops. (Summary by Mae Mercado) The Plant Cell 10.1093/plcell/koaf019
From water to land: What bryophytes reveal about plant evolution and adaptations
 The transition of plants from aquatic to terrestrial environments occurred approximately 400 million years ago, leading to the diversification of two major lineages: tracheophytes (vascular plants) and bryophytes (non-vascular plants). While most studies on plant adaptation to environmental stressors focus on angiosperms— which include the majority of crop species—similar research in bryophytes remains less explored. However, bryophytes provide insights into the characteristics of the most recent common ancestor of land plants. Using a pangenome approach, Beaulieu and colleagues conducted a genome-environment association study on 133 accessions of the bryophyte Marchantia. Their analyses revealed conserved molecular mechanisms across land plants, with nucleotide-binding leucine-rich repeat (NLR) gene families and class III peroxidases emerging as key adaptive strategies. These findings demonstrate that bryophyte studies can complement and inform research on angiosperms. Intraspecific diversity in bryophytes was also evident, particularly in genes involved in the biosynthesis and storage of oil bodies, which stood out as lineage-specific determinants and may have evolved through horizontal gene transfer. This extensive dataset provides a valuable resource for future studies to uncover new regulators of plant-environment interactions across land plant species. (Summary by Ching Chan @ntnuchanlab) Nature Genetics 10.1038/s41588-024-02071-4
The transition of plants from aquatic to terrestrial environments occurred approximately 400 million years ago, leading to the diversification of two major lineages: tracheophytes (vascular plants) and bryophytes (non-vascular plants). While most studies on plant adaptation to environmental stressors focus on angiosperms— which include the majority of crop species—similar research in bryophytes remains less explored. However, bryophytes provide insights into the characteristics of the most recent common ancestor of land plants. Using a pangenome approach, Beaulieu and colleagues conducted a genome-environment association study on 133 accessions of the bryophyte Marchantia. Their analyses revealed conserved molecular mechanisms across land plants, with nucleotide-binding leucine-rich repeat (NLR) gene families and class III peroxidases emerging as key adaptive strategies. These findings demonstrate that bryophyte studies can complement and inform research on angiosperms. Intraspecific diversity in bryophytes was also evident, particularly in genes involved in the biosynthesis and storage of oil bodies, which stood out as lineage-specific determinants and may have evolved through horizontal gene transfer. This extensive dataset provides a valuable resource for future studies to uncover new regulators of plant-environment interactions across land plant species. (Summary by Ching Chan @ntnuchanlab) Nature Genetics 10.1038/s41588-024-02071-4
Two blue-light photoreceptors (cry1 and phot1) function differently in hypocotyl light responses
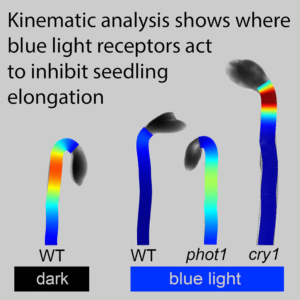 Plants have several types of light receptors that control various responses to light, such as leaf expansion, time-of-flowering, and hypocotyl elongation. When seeds are germinated in the dark, their hypocotyl elongates rapidly because darkness is perceived as being deep underground; the elongation is an effort to reach light. Hypocotyl elongation is quickly arrested in the presence of blue light. In this new work, Bustamante et al. developed a sophisticated approach to monitor hypocotyl responses to blue light in wild type plants as well as loss-of-function mutants of two blue light receptors, cry1 and phot1. Previous studies showed that although blue-light slows hypocotyl elongation in cry1 mutants, that effect is transient, and the mutant hypocotyls resume elongation after a short period. The authors set out to identify why the cry1 hypocotyls transiently pause elongation, and how different cells in the hypocotyl respond to light signals. The authors discuss several of the challenges in precisely determinig the relative elemental growth rate (REGR) in various parts of the Arabidopsis hypocotyl. Because measuring growth over time requires a non-invasive procedure, cell length must be determined in intact hypocotyls, which is complicated by the fact that the hypocotyl twists and bends. The authors therefore used machine learning to develop a pipeline for analyzing the images, which included identifying a hypocotyl midline for reference (see the paper for details). The results show that phot1 is required for blue light-induced elongation suppression, and that cry1-suppressed elongation occurs in a more apical hypocotyl region that was not previously known to be involved in the elongation response. This paper is an elegant example of how machine learning can be employed for high-resolution data analysis that can reveal novel mechanisms of plant development. (Summary by Mary Williams @PlantTeaching.bsky.social) Curr. Biol. 10.1016/j.cub.2024.11.021
Plants have several types of light receptors that control various responses to light, such as leaf expansion, time-of-flowering, and hypocotyl elongation. When seeds are germinated in the dark, their hypocotyl elongates rapidly because darkness is perceived as being deep underground; the elongation is an effort to reach light. Hypocotyl elongation is quickly arrested in the presence of blue light. In this new work, Bustamante et al. developed a sophisticated approach to monitor hypocotyl responses to blue light in wild type plants as well as loss-of-function mutants of two blue light receptors, cry1 and phot1. Previous studies showed that although blue-light slows hypocotyl elongation in cry1 mutants, that effect is transient, and the mutant hypocotyls resume elongation after a short period. The authors set out to identify why the cry1 hypocotyls transiently pause elongation, and how different cells in the hypocotyl respond to light signals. The authors discuss several of the challenges in precisely determinig the relative elemental growth rate (REGR) in various parts of the Arabidopsis hypocotyl. Because measuring growth over time requires a non-invasive procedure, cell length must be determined in intact hypocotyls, which is complicated by the fact that the hypocotyl twists and bends. The authors therefore used machine learning to develop a pipeline for analyzing the images, which included identifying a hypocotyl midline for reference (see the paper for details). The results show that phot1 is required for blue light-induced elongation suppression, and that cry1-suppressed elongation occurs in a more apical hypocotyl region that was not previously known to be involved in the elongation response. This paper is an elegant example of how machine learning can be employed for high-resolution data analysis that can reveal novel mechanisms of plant development. (Summary by Mary Williams @PlantTeaching.bsky.social) Curr. Biol. 10.1016/j.cub.2024.11.021
Plasmodesmata: A new frontier for membrane contact site intercellular communication
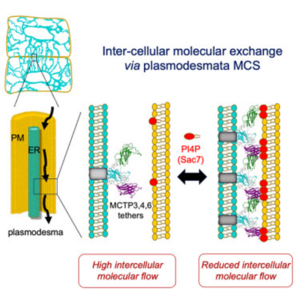 Membrane contact sites (MCSs) serve a critical role in intracellular communication, particularly between organelle membranes, enabling direct molecular transfer. Less clear is the role MCSs play in intercellular communication. New evidence from Pérez-Sancho et al. implicates plasmodesmata in this function and their endoplasmic reticulum-plasma membrane (ER-PM) MCSs. Unique to land plants and some green algae is an ER network that spans between cells (which is closely associated with the PM to form plasmodesmata). Historically, callose synthesis/degradation was the only established process for understanding plasmodesmata trafficking flow. The authors hypothesized that the MCS within plasmodesmata might act as a control valve for intercellular communication. They first identified core ER-PM tethering proteins (MCTP3, MCTP4, and MCTP6), then observed greater plasmodesmata diameter in Arabidopsis thaliana mutants for these proteins. Further, the mutants had faster molecular flow and reduced dynamic control when exposed to stressors compared to wild type, all independent of callose deposition mechanisms. The authors also characterized an anionic phospholipid (PI4P) that regulates MCTP4 docking to the PM, collectively working to fluctuate plasmodesmata permeability highly dependent on their own molecular regulators (including SAC7), which were shown to be differentially expressed in root cell types. Taken together, Pérez-Sancho et al. have highlighted an intricate regulatory network between cells via plasmodesmata. Their work finds a new intercellular function for MCSs with a control mechanism beyond just callose deposition. (Summary by Stephanie Temnyk @STemnyk) Cell 10.1016/j.cell.2024.11.034
Membrane contact sites (MCSs) serve a critical role in intracellular communication, particularly between organelle membranes, enabling direct molecular transfer. Less clear is the role MCSs play in intercellular communication. New evidence from Pérez-Sancho et al. implicates plasmodesmata in this function and their endoplasmic reticulum-plasma membrane (ER-PM) MCSs. Unique to land plants and some green algae is an ER network that spans between cells (which is closely associated with the PM to form plasmodesmata). Historically, callose synthesis/degradation was the only established process for understanding plasmodesmata trafficking flow. The authors hypothesized that the MCS within plasmodesmata might act as a control valve for intercellular communication. They first identified core ER-PM tethering proteins (MCTP3, MCTP4, and MCTP6), then observed greater plasmodesmata diameter in Arabidopsis thaliana mutants for these proteins. Further, the mutants had faster molecular flow and reduced dynamic control when exposed to stressors compared to wild type, all independent of callose deposition mechanisms. The authors also characterized an anionic phospholipid (PI4P) that regulates MCTP4 docking to the PM, collectively working to fluctuate plasmodesmata permeability highly dependent on their own molecular regulators (including SAC7), which were shown to be differentially expressed in root cell types. Taken together, Pérez-Sancho et al. have highlighted an intricate regulatory network between cells via plasmodesmata. Their work finds a new intercellular function for MCSs with a control mechanism beyond just callose deposition. (Summary by Stephanie Temnyk @STemnyk) Cell 10.1016/j.cell.2024.11.034
Developmental robustness from antagonizing cis-elements
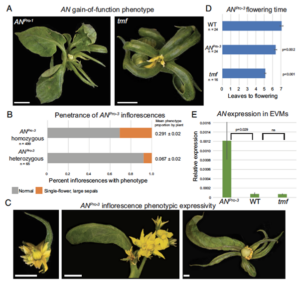 Developmental transitions in plants are tightly regulated by transcriptional networks that require fine-tuned temporal and spatial control, with noncoding sequences in gene promoters playing a key role. The conserved transcriptional regulator UNUSUAL FLORAL ORGANS (UFO) is essential for floral development. In this study, Lanctot et al. identified highly conserved sequences within the UFO promoter in tomato and Arabidopsis, highlighting their role in cis-regulation. Focusing first on tomato, the authors analyzed the promoter of the UFO ortholog, ANANTHA (AN). They identified conserved noncoding sequences (CNSs) and a chromatin accessibility “hotspot” indicative of cis-regulatory activity. CRISPR editing of this region caused strong phenotypic alterations in inflorescence and floral development. A gain-of-function mutant revealed that the deletion of a transcription factor binding site is associated with AN gain of function, with variability in expressivity suggesting the involvement of both activators and repressors. Biallelic mutant analysis confirmed that a balanced dosage of these factors is crucial for robust floral architecture. Extending their analysis to Arabidopsis, the authors found that while homologous regulatory sequences are more dispersed within the UFO promoter, they maintain similar functional control. However, phenotypic differences in mutant lines suggest species-specific regulatory divergence due to differing UFO expression patterns. This study highlights the complexity of cis-regulatory mechanisms in dynamic developmental genes, demonstrating that short, conserved sequences recruit both activators and repressors to maintain developmental robustness across species. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) PNAS, 10.1073/pnas.2421990122
Developmental transitions in plants are tightly regulated by transcriptional networks that require fine-tuned temporal and spatial control, with noncoding sequences in gene promoters playing a key role. The conserved transcriptional regulator UNUSUAL FLORAL ORGANS (UFO) is essential for floral development. In this study, Lanctot et al. identified highly conserved sequences within the UFO promoter in tomato and Arabidopsis, highlighting their role in cis-regulation. Focusing first on tomato, the authors analyzed the promoter of the UFO ortholog, ANANTHA (AN). They identified conserved noncoding sequences (CNSs) and a chromatin accessibility “hotspot” indicative of cis-regulatory activity. CRISPR editing of this region caused strong phenotypic alterations in inflorescence and floral development. A gain-of-function mutant revealed that the deletion of a transcription factor binding site is associated with AN gain of function, with variability in expressivity suggesting the involvement of both activators and repressors. Biallelic mutant analysis confirmed that a balanced dosage of these factors is crucial for robust floral architecture. Extending their analysis to Arabidopsis, the authors found that while homologous regulatory sequences are more dispersed within the UFO promoter, they maintain similar functional control. However, phenotypic differences in mutant lines suggest species-specific regulatory divergence due to differing UFO expression patterns. This study highlights the complexity of cis-regulatory mechanisms in dynamic developmental genes, demonstrating that short, conserved sequences recruit both activators and repressors to maintain developmental robustness across species. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) PNAS, 10.1073/pnas.2421990122
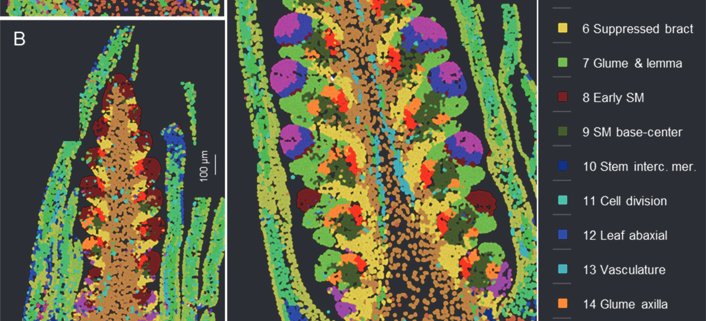
Single cell analysis of wheat spike development
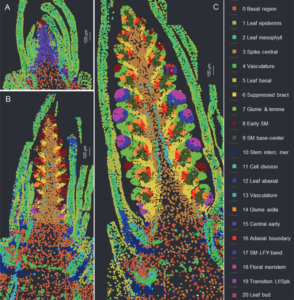 This preprint wins the award for “most beautiful paper” this week. Xu, Lin et al. carried out an expression analysis of developing wheat spikes at three developmental stages, using both single-cell RNA sequencing and single-molecule fluorescence in situ hybridization (smFISH). The wheat spike is the inflorescence where the grain ultimately forms, so it is responsible for producing a large proportion of human calories. The authors clustered genes by expression patterns and mapped some of them onto the tissue using smFISH. This enabled them to identify how gene expression patterns change during the development of the spikelet, and also to identify candidate genes that might control the size of the spike and number of flowers it could potentially form, opening the door to developing wheat with more grain per spike. There’s a wealth of information here and in the supplemental data, so if you’re interested in reproductive development you can see where your favorite genes are expressed in wheat. Everyone else can simply enjoy looking at these gorgeous images. (Summary by Mary Williams @PlantTeaching.bsky.social) bioRxiv https://doi.org/10.1101/2025.02.15.638402
This preprint wins the award for “most beautiful paper” this week. Xu, Lin et al. carried out an expression analysis of developing wheat spikes at three developmental stages, using both single-cell RNA sequencing and single-molecule fluorescence in situ hybridization (smFISH). The wheat spike is the inflorescence where the grain ultimately forms, so it is responsible for producing a large proportion of human calories. The authors clustered genes by expression patterns and mapped some of them onto the tissue using smFISH. This enabled them to identify how gene expression patterns change during the development of the spikelet, and also to identify candidate genes that might control the size of the spike and number of flowers it could potentially form, opening the door to developing wheat with more grain per spike. There’s a wealth of information here and in the supplemental data, so if you’re interested in reproductive development you can see where your favorite genes are expressed in wheat. Everyone else can simply enjoy looking at these gorgeous images. (Summary by Mary Williams @PlantTeaching.bsky.social) bioRxiv https://doi.org/10.1101/2025.02.15.638402
A mutation in a nuclear-membrane localized calcium channel enhances symbiosis
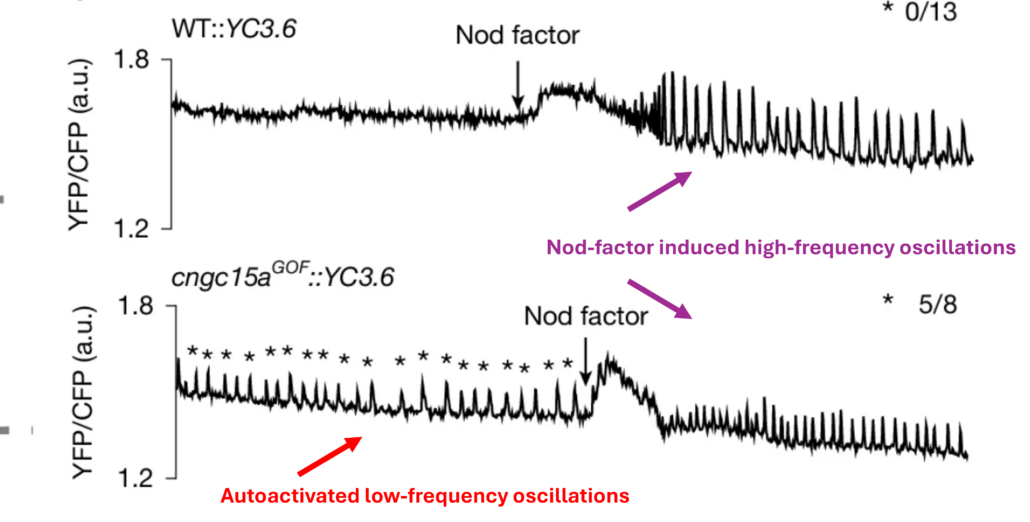 Calcium oscillations are widely employed molecular signals, but how signals are encoded and decoded remains largely unknown. In a new study, Cook et al. investigated mutations in the nuclear envelope-localized calcium channel CNGC15 from the legume Medicago truncatula. This protein normally forms a complex with a second calcium channel protein, DMI1, and together they mediate signaling between plants and endosymbiotic fungi and bacteria and induce symbiosis genes in the presence of the symbiont. The authors identified a single-amino acid mutation of CNGC15 that enables it to produce calcium oscillations independently of DMI1 and Nod-factors (signals from rhizobia symbionts); the authors describe the product of the gain-of-function mutation (CNGC15GOF) as an autoactivated channel. Interestingly, the frequency of calcium oscillations in the GOF mutant was less than that of that of the wild-type channel. However, in the presence of DMI1 and the Nod-factor signal, the frequency was restored to its wild-type rate; thus, CNGC15GOF produces two types of signals, the autoactivated low-frequency signal and the endosymbiont-activated high-frequency signal. Although symbiosis genes were not expressed in the CNGC15GOF plants in the absence of the symbionts, the plants did increase the production of flavonoids, which serve as attractants to symbionts. Furthermore, the plants expressing CNGC15GOF showed enhanced symbiosis with both arbuscular mycorrhiza and rhizobia. Finally, the authors identified wheat lines with the corresponding channel mutation and found that these lines showed significantly enhanced AM colonization, both in controlled and field conditions. These results show the low-frequency calcium oscillations generated by the autoactivated channel promote symbiosis, raising the possibility of lowering fertilizer used without losing crop productivity. (Summary by Mary Williams @PlantTeaching.bsky.social) Nature 10.1038/s41586-024-08424-7
Calcium oscillations are widely employed molecular signals, but how signals are encoded and decoded remains largely unknown. In a new study, Cook et al. investigated mutations in the nuclear envelope-localized calcium channel CNGC15 from the legume Medicago truncatula. This protein normally forms a complex with a second calcium channel protein, DMI1, and together they mediate signaling between plants and endosymbiotic fungi and bacteria and induce symbiosis genes in the presence of the symbiont. The authors identified a single-amino acid mutation of CNGC15 that enables it to produce calcium oscillations independently of DMI1 and Nod-factors (signals from rhizobia symbionts); the authors describe the product of the gain-of-function mutation (CNGC15GOF) as an autoactivated channel. Interestingly, the frequency of calcium oscillations in the GOF mutant was less than that of that of the wild-type channel. However, in the presence of DMI1 and the Nod-factor signal, the frequency was restored to its wild-type rate; thus, CNGC15GOF produces two types of signals, the autoactivated low-frequency signal and the endosymbiont-activated high-frequency signal. Although symbiosis genes were not expressed in the CNGC15GOF plants in the absence of the symbionts, the plants did increase the production of flavonoids, which serve as attractants to symbionts. Furthermore, the plants expressing CNGC15GOF showed enhanced symbiosis with both arbuscular mycorrhiza and rhizobia. Finally, the authors identified wheat lines with the corresponding channel mutation and found that these lines showed significantly enhanced AM colonization, both in controlled and field conditions. These results show the low-frequency calcium oscillations generated by the autoactivated channel promote symbiosis, raising the possibility of lowering fertilizer used without losing crop productivity. (Summary by Mary Williams @PlantTeaching.bsky.social) Nature 10.1038/s41586-024-08424-7
Fungal pathogen hijacks phosphate signaling to promote pathogenicity
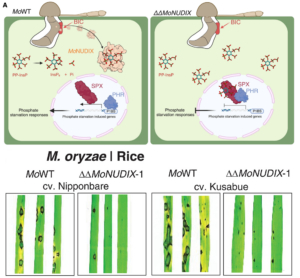 Weakening your opponent is an effective way to win a battle. A new paper by McCombe et al. shows that several pathogenic fungi weaken their plant hosts by disrupting their phosphate signaling pathways. Phosphate is indispensable for plant growth, as a component of ATP, nucleotides, phospholipids, and countless small molecules, so plants have complex systems for taking up and monitoring phosphate. Inositol pyrophosphate (PP-InsP) is a small, phosphate-rich molecule that serves as a measure of P sufficiency by interacting with key regulators to suppress the phosphate starvation response. McCombe et al. studied a class of effector proteins secreted by plant pathogens called Nudix (nucleoside-diphosphate linked to moiety X) proteins and found that they hydrolyze PP-InsP. Nudix genes are highly upregulated upon infection and knocking them out severely limits pathogen virulence. The authors initially looked at the interaction between Magnaporthe oryzae and rice, but found that Nudix effectors have similar properties in other pathogenic fungi (Colletotrichum spp.) acting on other plants. The authors confirmed that the PP-InsP hydrolase activity is required for the disease promotion by the Nudix proteins, and that they hydrolyze PP-InsP in plant cells and promote the starvation response, which may be sufficient to weaken the plant as it unnecessarily allocates energy to this pathway. However, the authors observe that the pathogen might also benefit nutritionally from the elevated intercellular P as the plant naively accumulates P. Furthermore, PP-InsPs have a role in activating jasmonate signaling, so lowering PP-InsP levels might additionally benefit the pathogen through suppressed defense signaling. This is a fascinating study showing once again that all is NOT fair in love and war. Have a look also at the accompanying Perspective by Caroline Gutjahr 10.1126/science.adw1568. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adl5764
Weakening your opponent is an effective way to win a battle. A new paper by McCombe et al. shows that several pathogenic fungi weaken their plant hosts by disrupting their phosphate signaling pathways. Phosphate is indispensable for plant growth, as a component of ATP, nucleotides, phospholipids, and countless small molecules, so plants have complex systems for taking up and monitoring phosphate. Inositol pyrophosphate (PP-InsP) is a small, phosphate-rich molecule that serves as a measure of P sufficiency by interacting with key regulators to suppress the phosphate starvation response. McCombe et al. studied a class of effector proteins secreted by plant pathogens called Nudix (nucleoside-diphosphate linked to moiety X) proteins and found that they hydrolyze PP-InsP. Nudix genes are highly upregulated upon infection and knocking them out severely limits pathogen virulence. The authors initially looked at the interaction between Magnaporthe oryzae and rice, but found that Nudix effectors have similar properties in other pathogenic fungi (Colletotrichum spp.) acting on other plants. The authors confirmed that the PP-InsP hydrolase activity is required for the disease promotion by the Nudix proteins, and that they hydrolyze PP-InsP in plant cells and promote the starvation response, which may be sufficient to weaken the plant as it unnecessarily allocates energy to this pathway. However, the authors observe that the pathogen might also benefit nutritionally from the elevated intercellular P as the plant naively accumulates P. Furthermore, PP-InsPs have a role in activating jasmonate signaling, so lowering PP-InsP levels might additionally benefit the pathogen through suppressed defense signaling. This is a fascinating study showing once again that all is NOT fair in love and war. Have a look also at the accompanying Perspective by Caroline Gutjahr 10.1126/science.adw1568. (Summary by Mary Williams @PlantTeaching.bsky.social) Science 10.1126/science.adl5764
Blocking bacterial invasion: Erucamide inhibits the type III secretion system
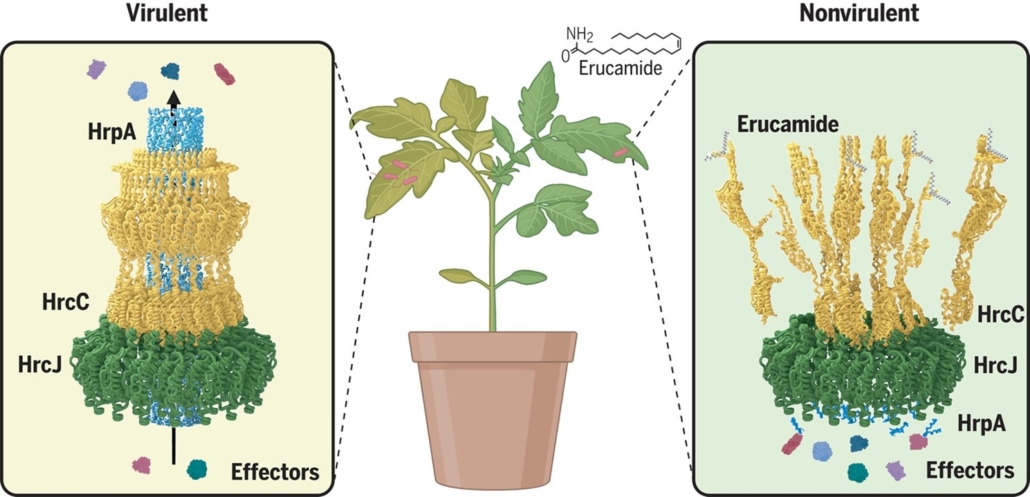 The interaction between plants and pathogens is often described as an evolutionary arms race, in which plants develop multilayered immune signaling to counteract direct extracellular attacks and intracellular chemical effectors. The type III secretion system (T3SS) of Gram-negative bacteria serves as a key weapon, allowing pathogens to inject effectors directly into plant cells to suppress immune responses and promote infection. Although it is widely accepted that T3SS can be inhibited by plant immune signaling, the key determinants of this inhibition remain unknown. Miao and colleagues hypothesized that certain secondary metabolites may play a role in this inhibition. Using activity-guided chemical purification, they tested metabolites extracted through multiple rounds of organic solvent fractionation for their ability to inhibit secretion of the bacterial effector AvrPto in vitro. Notably, a methylene chloride fraction exhibited a strong inhibitory effect, and by using nuclear magnetic resonance and high-resolution mass spectrometry the active compound was identified as erucamide, a plant phytoalexin. Further investigation through homology modeling and mutant studies confirmed the specificity of this interaction; the authors speculate that erucamide might disrupt the assembly of the T3SS. This study clarifies the long-sought-after inhibitor of T3SS, highlights the critical function of erucamide, and lays the foundation for the development of novel pesticide strategies. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.ads0377
The interaction between plants and pathogens is often described as an evolutionary arms race, in which plants develop multilayered immune signaling to counteract direct extracellular attacks and intracellular chemical effectors. The type III secretion system (T3SS) of Gram-negative bacteria serves as a key weapon, allowing pathogens to inject effectors directly into plant cells to suppress immune responses and promote infection. Although it is widely accepted that T3SS can be inhibited by plant immune signaling, the key determinants of this inhibition remain unknown. Miao and colleagues hypothesized that certain secondary metabolites may play a role in this inhibition. Using activity-guided chemical purification, they tested metabolites extracted through multiple rounds of organic solvent fractionation for their ability to inhibit secretion of the bacterial effector AvrPto in vitro. Notably, a methylene chloride fraction exhibited a strong inhibitory effect, and by using nuclear magnetic resonance and high-resolution mass spectrometry the active compound was identified as erucamide, a plant phytoalexin. Further investigation through homology modeling and mutant studies confirmed the specificity of this interaction; the authors speculate that erucamide might disrupt the assembly of the T3SS. This study clarifies the long-sought-after inhibitor of T3SS, highlights the critical function of erucamide, and lays the foundation for the development of novel pesticide strategies. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.ads0377
Stealth mode: How Rhodanobacter R179 evades plant immunity
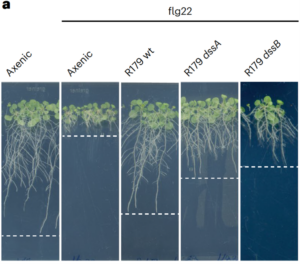 The soil microbiome harbors a vast diversity of microorganisms that can be pathogenic, beneficial, or commensal to plants. A fundamental question in plant biology is how plants actively detect, differentiate, and optimize their associations with the microbiome to maintain optimal fitness. In a recent study, Ordon and colleagues took an alternative approach—exploring how bacteria evade plant immune surveillance. The bacterial families Xanthomonadaceae and Rhodanobacteraceae include both commensal and pathogenic strains. Interestingly, Rhodanobacter R179, a root commensal belonging to a branch of Xanthomonadales, possesses immunogenic elicitors recognized by the plant immune receptors EFR and SOBIR1. However, it does not trigger a pattern-triggered immunity (PTI) response, such as flg22-induced root growth inhibition (RGI). By screening a collection of R179 mutants, the researchers identified two transporter complex mutants, dssA and dssB, which fail to mediate peptidase secretion and become sensitive to flg22-induced RGI. The ability to secrete peptidase allows R179 to eliminate immunogenic peptides and evade plant immunity. This effect extends beyond R179, as the secreted peptidase influences other microbiota members within the soil matrix. In a community context, R179 promotes microbiota diversity but at the cost of increased metabolic constraints on itself. The rationale and mechanism behind this coexistence and its ecological niche remain intriguing and warrant further investigation. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-025-01918-w
The soil microbiome harbors a vast diversity of microorganisms that can be pathogenic, beneficial, or commensal to plants. A fundamental question in plant biology is how plants actively detect, differentiate, and optimize their associations with the microbiome to maintain optimal fitness. In a recent study, Ordon and colleagues took an alternative approach—exploring how bacteria evade plant immune surveillance. The bacterial families Xanthomonadaceae and Rhodanobacteraceae include both commensal and pathogenic strains. Interestingly, Rhodanobacter R179, a root commensal belonging to a branch of Xanthomonadales, possesses immunogenic elicitors recognized by the plant immune receptors EFR and SOBIR1. However, it does not trigger a pattern-triggered immunity (PTI) response, such as flg22-induced root growth inhibition (RGI). By screening a collection of R179 mutants, the researchers identified two transporter complex mutants, dssA and dssB, which fail to mediate peptidase secretion and become sensitive to flg22-induced RGI. The ability to secrete peptidase allows R179 to eliminate immunogenic peptides and evade plant immunity. This effect extends beyond R179, as the secreted peptidase influences other microbiota members within the soil matrix. In a community context, R179 promotes microbiota diversity but at the cost of increased metabolic constraints on itself. The rationale and mechanism behind this coexistence and its ecological niche remain intriguing and warrant further investigation. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-025-01918-w