Plant Science Research Weekly: March 29, 2024
Review. Chloroplast ATP synthase: From structure to engineering
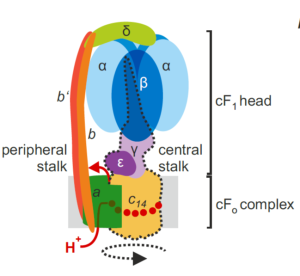 I remember how amazed I was the first time I saw an animation of ATP synthase doing its job. This fantastic engine is largely conserved across the domains of life, with some variation as highlighted in this review of the chloroplast ATP synthase by Rühle et al. The plastid form is a rotary form, written as cF1F0. The cF0 subunit is embedded in the thylakoid membrane, whereas the cF1 subunit protrudes into the stroma. Protons move through it (driven by proton-motive force generated by photosynthesis) from within the lumen across the membrane into the stroma, and their movement is coupled with the phosphorylation of ADP to ATP. The review is divided into three parts: structure, assembly, and engineering, and I particularly enjoyed the discussion of how the chloroplast enzyme is regulated by redox, pH, and in turn regulates the light reactions including the balance between cyclic and linear electron flow and PSII and PSI activities. Because of the intricate connection between protons, electrons, light reactions and ATP, efforts to make the ATPase more efficient are challenging but intriguing. However, given the many efforts to improve other components of photosynthetic machinery, such as the introduction of carbon-concentrating mechanisms, understanding how such efforts might need to also be supported through engineering the chloroplast ATPase is critical. Note that this review is part of an upcoming Plant Cell Focus Issue on Photosynthesis. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae081
I remember how amazed I was the first time I saw an animation of ATP synthase doing its job. This fantastic engine is largely conserved across the domains of life, with some variation as highlighted in this review of the chloroplast ATP synthase by Rühle et al. The plastid form is a rotary form, written as cF1F0. The cF0 subunit is embedded in the thylakoid membrane, whereas the cF1 subunit protrudes into the stroma. Protons move through it (driven by proton-motive force generated by photosynthesis) from within the lumen across the membrane into the stroma, and their movement is coupled with the phosphorylation of ADP to ATP. The review is divided into three parts: structure, assembly, and engineering, and I particularly enjoyed the discussion of how the chloroplast enzyme is regulated by redox, pH, and in turn regulates the light reactions including the balance between cyclic and linear electron flow and PSII and PSI activities. Because of the intricate connection between protons, electrons, light reactions and ATP, efforts to make the ATPase more efficient are challenging but intriguing. However, given the many efforts to improve other components of photosynthetic machinery, such as the introduction of carbon-concentrating mechanisms, understanding how such efforts might need to also be supported through engineering the chloroplast ATPase is critical. Note that this review is part of an upcoming Plant Cell Focus Issue on Photosynthesis. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae081
Review: Evolution of ROS targets for plant development
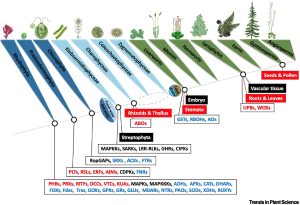 Reactive oxygen species (ROS) are agents of damage but also potent signals. Here, Singh et al. review the cellular targets that support ROS signaling across the green kingdom. Many of the signaling roles for ROS have been uncovered in Arabidopsis and other angiosperms, so it is interesting to look at their presence in other plant types. Most of the ROS signaling targets are present in green algae and thus predate terrestrialization, but there has also been a notable diversification of these gene families in terrestrial plants. The article has a particular focus on the roles of ROS in root and leaf developmental programs, and how they interact with phytohormones. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.03.005
Reactive oxygen species (ROS) are agents of damage but also potent signals. Here, Singh et al. review the cellular targets that support ROS signaling across the green kingdom. Many of the signaling roles for ROS have been uncovered in Arabidopsis and other angiosperms, so it is interesting to look at their presence in other plant types. Most of the ROS signaling targets are present in green algae and thus predate terrestrialization, but there has also been a notable diversification of these gene families in terrestrial plants. The article has a particular focus on the roles of ROS in root and leaf developmental programs, and how they interact with phytohormones. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.03.005
Review. From stressed to success: Unveiling the secret memory of gymnosperms
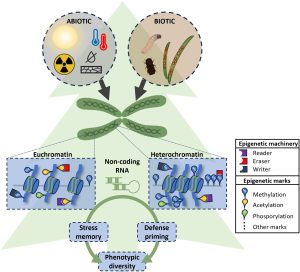 Epigenetic stress memory in gymnosperms is the topic of a new review by Fossdal et al. Gymnosperms are long-lived, cone-bearing, tough trees, known to tolerate episodic stress. Their adaptive traits to stress are not passed on in a straightforward way, conforming to classical Mendelian inheritance. Instead, adaptive responses in gymnosperms are a result of epigenetic changes, including DNA methylation, histone modification, and small RNAs, which enable dynamic changes in gene expression, thus, phenotypic expression. As an example of a epigenetic-controlled trait, in Norway spruce tree (Picea abies) the epigenetic memory of photoperiod and temperature experienced during embryogenesis and seed development influences bud phenology and the progenies’ frost tolerance many years later. The authors also highlight that most of our understanding of epigenetic regulations in plants comes from angiosperms, flowering plants. Studying epigenetic regulation in gymnosperms is more challenging due to their long lifecycle, rendering patterns of inheritance difficult to observe. Furthermore, the woody structure of gymnosperms makes the use of conventional nucleotide extraction methods less effective, and the only existing high quality reference genome belongs to Chinese pine (Pinus tabuliformis). Therefore, Fossdal et al., emphasize the need of improved sequencing technologies to decipher the large genomes of gymnosperms. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Physiol. 10.1093/plphys/kiae051/7595554
Epigenetic stress memory in gymnosperms is the topic of a new review by Fossdal et al. Gymnosperms are long-lived, cone-bearing, tough trees, known to tolerate episodic stress. Their adaptive traits to stress are not passed on in a straightforward way, conforming to classical Mendelian inheritance. Instead, adaptive responses in gymnosperms are a result of epigenetic changes, including DNA methylation, histone modification, and small RNAs, which enable dynamic changes in gene expression, thus, phenotypic expression. As an example of a epigenetic-controlled trait, in Norway spruce tree (Picea abies) the epigenetic memory of photoperiod and temperature experienced during embryogenesis and seed development influences bud phenology and the progenies’ frost tolerance many years later. The authors also highlight that most of our understanding of epigenetic regulations in plants comes from angiosperms, flowering plants. Studying epigenetic regulation in gymnosperms is more challenging due to their long lifecycle, rendering patterns of inheritance difficult to observe. Furthermore, the woody structure of gymnosperms makes the use of conventional nucleotide extraction methods less effective, and the only existing high quality reference genome belongs to Chinese pine (Pinus tabuliformis). Therefore, Fossdal et al., emphasize the need of improved sequencing technologies to decipher the large genomes of gymnosperms. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Physiol. 10.1093/plphys/kiae051/7595554
Making genome editing a success story in Africa
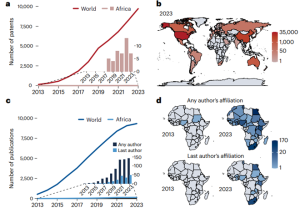 The introduction of CRISPR-Cas technology in 2012 marked a significant advancement in global genome editing, yet its potential remains largely untapped in Africa, where it could address crucial challenges in agriculture, public health, and medicine. However, several obstacles hinder its full realization, as addressed in this article by Abkallo et al. These include regulatory uncertainty, limited access to laboratories, equipment, reagents, and overall funding. Additionally, there is a shortage of skilled professionals and insufficient public support, resulting in a lack of political will for change. To overcome these challenges, various solutions have been proposed. These include establishing incentive programs to retain talent, increasing public awareness and education to facilitate informed decision-making, developing GMO regulatory frameworks and harmonizing them across the continent, and encouraging private investment. Furthermore, funding from global bodies such as the World Health Organization should be utilized to establish infrastructure suitable for genome-editing technologies. By implementing these measures, Africa can unlock the transformative potential of genome editing, fostering innovation and development throughout the continent. (Summary by Villő Bernád) Nature Biotech. 10.1038/s41587-024-02187-2
The introduction of CRISPR-Cas technology in 2012 marked a significant advancement in global genome editing, yet its potential remains largely untapped in Africa, where it could address crucial challenges in agriculture, public health, and medicine. However, several obstacles hinder its full realization, as addressed in this article by Abkallo et al. These include regulatory uncertainty, limited access to laboratories, equipment, reagents, and overall funding. Additionally, there is a shortage of skilled professionals and insufficient public support, resulting in a lack of political will for change. To overcome these challenges, various solutions have been proposed. These include establishing incentive programs to retain talent, increasing public awareness and education to facilitate informed decision-making, developing GMO regulatory frameworks and harmonizing them across the continent, and encouraging private investment. Furthermore, funding from global bodies such as the World Health Organization should be utilized to establish infrastructure suitable for genome-editing technologies. By implementing these measures, Africa can unlock the transformative potential of genome editing, fostering innovation and development throughout the continent. (Summary by Villő Bernád) Nature Biotech. 10.1038/s41587-024-02187-2
Viewpoint: Unheard voices from the Global South speak up
 For many reasons, the voices of researchers from the Global South often go unheard. Many of these reasons are financial; big international conferences are frequently held at places that require distant travel, and registration and travel fees can be beyond the reach of modestly-funded scientists. Visa challenges and systemic biases also contribute. In 2023, Auge and Estévez coordinated a concurrent symposium at the International Conference on Arabidopsis Research (ICAR) to highlight these unheard voices. In this article, the authors further amplify the voices of the symposium speakers but also the impediments to their being heard, and they provide recommendations to secure greater diversity in academic conferences. (Summary by Mary Williams @PlantTeaching) J. Exp. Bot. 10.1093/jxb/erae027
For many reasons, the voices of researchers from the Global South often go unheard. Many of these reasons are financial; big international conferences are frequently held at places that require distant travel, and registration and travel fees can be beyond the reach of modestly-funded scientists. Visa challenges and systemic biases also contribute. In 2023, Auge and Estévez coordinated a concurrent symposium at the International Conference on Arabidopsis Research (ICAR) to highlight these unheard voices. In this article, the authors further amplify the voices of the symposium speakers but also the impediments to their being heard, and they provide recommendations to secure greater diversity in academic conferences. (Summary by Mary Williams @PlantTeaching) J. Exp. Bot. 10.1093/jxb/erae027
Thylakoid membrane remodelling during the dark-to-light transition
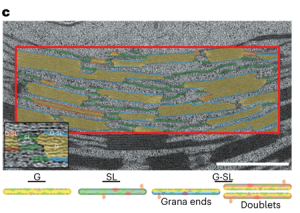 In the dark, plants modify thylakoid stacking to alter electron transport and reduce photodamage. More photosystem II (PSII) is located in thylakoids within stacked grana, which promotes cyclic electron transport. Upon light exposure, there is granal unstacking, which increases the amount of linear electron transport for carbon assimilation. Here Garty et al. used freeze fracture cryo scanning electron microscopy and transmission electron microscopy to study thylakoid membrane structure during the dark-to-light transition at high resolution. They categorized thylakoids in wild type Arabidopsis thaliana leaf chloroplasts into four groups – singular thylakoids exposed to the stroma on both sides, thylakoids within a granal stack, those on the edge of a granal stack and those in a thylakoid pair, which they called ‘stacked thylakoid doublets’. On the dark-to-light transition, the percentage of thylakoids in granal stacks decreased from 44% to 33%, whilst the percentage of stacked thylakoid doublets increased from 13% to 26%. Modelling these changes showed that the formation of stacked thylakoid doublets increased the fraction of PSII in proximity to PSI from 0.44 to 0.59, this allowed for more efficient linear electron transport. Hence, this study provides the first observations of stacked thylakoid doublets which are an ideal platform for linear electron transport. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-024-01628-9
In the dark, plants modify thylakoid stacking to alter electron transport and reduce photodamage. More photosystem II (PSII) is located in thylakoids within stacked grana, which promotes cyclic electron transport. Upon light exposure, there is granal unstacking, which increases the amount of linear electron transport for carbon assimilation. Here Garty et al. used freeze fracture cryo scanning electron microscopy and transmission electron microscopy to study thylakoid membrane structure during the dark-to-light transition at high resolution. They categorized thylakoids in wild type Arabidopsis thaliana leaf chloroplasts into four groups – singular thylakoids exposed to the stroma on both sides, thylakoids within a granal stack, those on the edge of a granal stack and those in a thylakoid pair, which they called ‘stacked thylakoid doublets’. On the dark-to-light transition, the percentage of thylakoids in granal stacks decreased from 44% to 33%, whilst the percentage of stacked thylakoid doublets increased from 13% to 26%. Modelling these changes showed that the formation of stacked thylakoid doublets increased the fraction of PSII in proximity to PSI from 0.44 to 0.59, this allowed for more efficient linear electron transport. Hence, this study provides the first observations of stacked thylakoid doublets which are an ideal platform for linear electron transport. (Summary by Rose McNelly @Rose_McN) Nature Plants 10.1038/s41477-024-01628-9
Lag, then leg it! An updated two-phase model for axillary bud activation and outgrowth in Arabidopsis
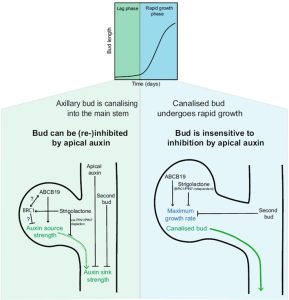 The mechanisms of apical dominance and bud outgrowth have puzzled generations of plant scientists, and over a century various hypotheses have been scrutinized. Two main hubs of regulation – auxin canalization (movement of auxin from a bud into the main stem) and the branching transcription factors BRANCHED 1/2 (BRC1/2) – act in parallel and are influenced by factors such as strigolactones, auxin-loading transporter ABCB19, and the activity of neighbouring buds. Here, Nahas et al. used one- and two-node Arabidopsis thaliana Col-0 inflorescence stem segments to examine bud activation and outgrowth and the contribution of external factors to the dynamics of branching. Within both intact and decapitated shoots, two populations of buds with different maximum growth rates (MGR) were found. The authors introduce a new two-phase developmental model from the data, comprising an activation phase (lag) followed by rapid growth. Buds in both intact and decapitated shoots have equivalent lag phase duration and MGRs. Apical application of auxin extends the lag phase for buds of decapitated shoots for up to 72 h; afterwards, buds become insensitive to it. When basally applied to decapitated shoots, a strigolactone analogue prolongs the lag phase and lowers MGRs. Neighbouring buds compete for access to the main stem auxin flux (canalization), prolonging the lag phase for both buds and lowering the MGR of the topmost bud. The two phases discussed herein are compatible with previously proposed models and seem to be genetically and physiologically dissociable. Upon decapitation, bud activation and its establishment as an auxin source, which is inhibited by strigolactones and BRC1/2, is helped by ABCB19. This leads to canalizing auxin transport to the stem, which is now an auxin sink. Once activated, the bud’s rapid outgrowth is unlikely to be reverted, but its growth rate is hampered by competing buds and strigolactone signaling. (Summary by Johnatan Vilasboa @vilasjohn) New Phytol. 10.1111/nph.19664
The mechanisms of apical dominance and bud outgrowth have puzzled generations of plant scientists, and over a century various hypotheses have been scrutinized. Two main hubs of regulation – auxin canalization (movement of auxin from a bud into the main stem) and the branching transcription factors BRANCHED 1/2 (BRC1/2) – act in parallel and are influenced by factors such as strigolactones, auxin-loading transporter ABCB19, and the activity of neighbouring buds. Here, Nahas et al. used one- and two-node Arabidopsis thaliana Col-0 inflorescence stem segments to examine bud activation and outgrowth and the contribution of external factors to the dynamics of branching. Within both intact and decapitated shoots, two populations of buds with different maximum growth rates (MGR) were found. The authors introduce a new two-phase developmental model from the data, comprising an activation phase (lag) followed by rapid growth. Buds in both intact and decapitated shoots have equivalent lag phase duration and MGRs. Apical application of auxin extends the lag phase for buds of decapitated shoots for up to 72 h; afterwards, buds become insensitive to it. When basally applied to decapitated shoots, a strigolactone analogue prolongs the lag phase and lowers MGRs. Neighbouring buds compete for access to the main stem auxin flux (canalization), prolonging the lag phase for both buds and lowering the MGR of the topmost bud. The two phases discussed herein are compatible with previously proposed models and seem to be genetically and physiologically dissociable. Upon decapitation, bud activation and its establishment as an auxin source, which is inhibited by strigolactones and BRC1/2, is helped by ABCB19. This leads to canalizing auxin transport to the stem, which is now an auxin sink. Once activated, the bud’s rapid outgrowth is unlikely to be reverted, but its growth rate is hampered by competing buds and strigolactone signaling. (Summary by Johnatan Vilasboa @vilasjohn) New Phytol. 10.1111/nph.19664
Neo-polyploid infertility associated with defective pollen tube growth
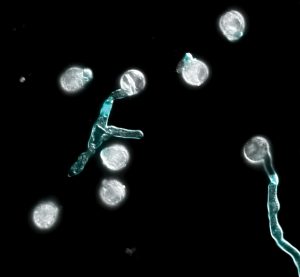 Polyploidy, arising from whole-genome duplication, is a common phenomenon in plant species. However, newly formed polyploids are often infertile and the mechanism(s) by which they adapt to their new karyotypes remain unclear. In this article, Westermann and colleagues chemically induced the formation of tetraploid Arabidopsis arenosa plants from diploid parents. They found that the lower fertility of these newly established tetraploids was due to physiological defects in pollen tube growth. They focused on two genes, AGC1.5 and ACA8, which are under selection in naturally occuring tetraploid populations of A. arenosa. Compared to their diploid counterparts, the majority of the established tetraploids, which have diploid-like fertility levels, exhibited specific allelic variants that differed by only one (AGC1.5) or two (ACA8) amino acids in their coding sequences, respectively. The genetic and phenotypic analysis of the F2 individiduals obtained by crossing the newly formed tetraploids with the established tetraploids showed that pollen tube growth was rescued in the individuals homozygus for the tetraploid alleles, while heterozygus individuals had an intermediate phenotype. This work is an example of the “reverse adaptation genomic approach”, starting from a set of candidate genes under selection and tracing back their contribution to plant fitness at the molecular level. It also offers new insights as to why new polyploids are often infertile and a possible way to improve this trait. (Summary by Carlo Pasini @Crl_Psn) Science 10.1126/science.adh0755
Polyploidy, arising from whole-genome duplication, is a common phenomenon in plant species. However, newly formed polyploids are often infertile and the mechanism(s) by which they adapt to their new karyotypes remain unclear. In this article, Westermann and colleagues chemically induced the formation of tetraploid Arabidopsis arenosa plants from diploid parents. They found that the lower fertility of these newly established tetraploids was due to physiological defects in pollen tube growth. They focused on two genes, AGC1.5 and ACA8, which are under selection in naturally occuring tetraploid populations of A. arenosa. Compared to their diploid counterparts, the majority of the established tetraploids, which have diploid-like fertility levels, exhibited specific allelic variants that differed by only one (AGC1.5) or two (ACA8) amino acids in their coding sequences, respectively. The genetic and phenotypic analysis of the F2 individiduals obtained by crossing the newly formed tetraploids with the established tetraploids showed that pollen tube growth was rescued in the individuals homozygus for the tetraploid alleles, while heterozygus individuals had an intermediate phenotype. This work is an example of the “reverse adaptation genomic approach”, starting from a set of candidate genes under selection and tracing back their contribution to plant fitness at the molecular level. It also offers new insights as to why new polyploids are often infertile and a possible way to improve this trait. (Summary by Carlo Pasini @Crl_Psn) Science 10.1126/science.adh0755
Herbivore-deterring trichomes persist with the help of Woolly and Get02
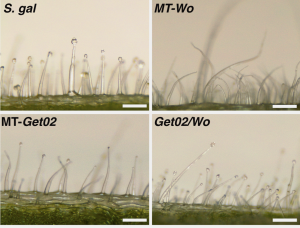 Type-IV glandular trichomes, which produce acylsugars, are effective deterrents against herbivory in Solanum, but they only persist in the juvenile stage of the cultivated tomato (S. lycopersicum). Therefore, these trichomes serve as a marker for the transition from juvenile to adult phases in developmental studies, as previously discovered by Vendemiatti et al. (2017). Now, Vendemiatti et al. have taken two strategies to promote acylsugar-based herbivory resistance in tomato. First, they mapped and introgressed a key locus from a tomato wild relative, S. galapagense that controls the capacity to harbor adult-persisting type-IV trichomes. In the resulting plants, MT-Get02 (Micro-Tom-Galapagos enhanced trichome 02), these trichomes are retained into the adult phase. However, the MT-Get02 plants only display about half as many type-IV trichomes as the parental S. galapagense plants. In S. lycopersicum, a mutation in the gene WOOLLY (Wom) leads to a considerable increase in the number of trichomes. The authors stacked the Wom mutation onto MT-Get02 and achieved plants with a high density of type-IV trichomes in both juvenile and adult phases, compared to the wild species. Interestingly, S. galapagense´s acyl-sugar composition differed from that of Get02/Wo plants, yet their quantities were similar, despite the lower density of type-IV in the wild species. This suggests the presence of additional factors influencing insect resistance in S. galapagense, such as specific acylsugar biosynthesis pathways and the capacity of acylsugar production per gland. Moreover, the combination of just two loci was sufficient to confer greater herbivory resistance to Get02/Wo plants compared to the S. lycopersicum parent, making it a significant breakthrough in overcoming the polygenic nature of insect resistance in tomatoes. And if you’d like to read more on Woolly’s role in trichome development, see the recently accepted The Plant Cell paper, “A gradient of the HD-Zip regulator Woolly regulates multicellular trichome morphogenesis in tomato” 10.1093/plcell/koae077. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae128
Type-IV glandular trichomes, which produce acylsugars, are effective deterrents against herbivory in Solanum, but they only persist in the juvenile stage of the cultivated tomato (S. lycopersicum). Therefore, these trichomes serve as a marker for the transition from juvenile to adult phases in developmental studies, as previously discovered by Vendemiatti et al. (2017). Now, Vendemiatti et al. have taken two strategies to promote acylsugar-based herbivory resistance in tomato. First, they mapped and introgressed a key locus from a tomato wild relative, S. galapagense that controls the capacity to harbor adult-persisting type-IV trichomes. In the resulting plants, MT-Get02 (Micro-Tom-Galapagos enhanced trichome 02), these trichomes are retained into the adult phase. However, the MT-Get02 plants only display about half as many type-IV trichomes as the parental S. galapagense plants. In S. lycopersicum, a mutation in the gene WOOLLY (Wom) leads to a considerable increase in the number of trichomes. The authors stacked the Wom mutation onto MT-Get02 and achieved plants with a high density of type-IV trichomes in both juvenile and adult phases, compared to the wild species. Interestingly, S. galapagense´s acyl-sugar composition differed from that of Get02/Wo plants, yet their quantities were similar, despite the lower density of type-IV in the wild species. This suggests the presence of additional factors influencing insect resistance in S. galapagense, such as specific acylsugar biosynthesis pathways and the capacity of acylsugar production per gland. Moreover, the combination of just two loci was sufficient to confer greater herbivory resistance to Get02/Wo plants compared to the S. lycopersicum parent, making it a significant breakthrough in overcoming the polygenic nature of insect resistance in tomatoes. And if you’d like to read more on Woolly’s role in trichome development, see the recently accepted The Plant Cell paper, “A gradient of the HD-Zip regulator Woolly regulates multicellular trichome morphogenesis in tomato” 10.1093/plcell/koae077. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae128
An inducible protein degradation system for rapid depletion of target proteins
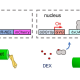
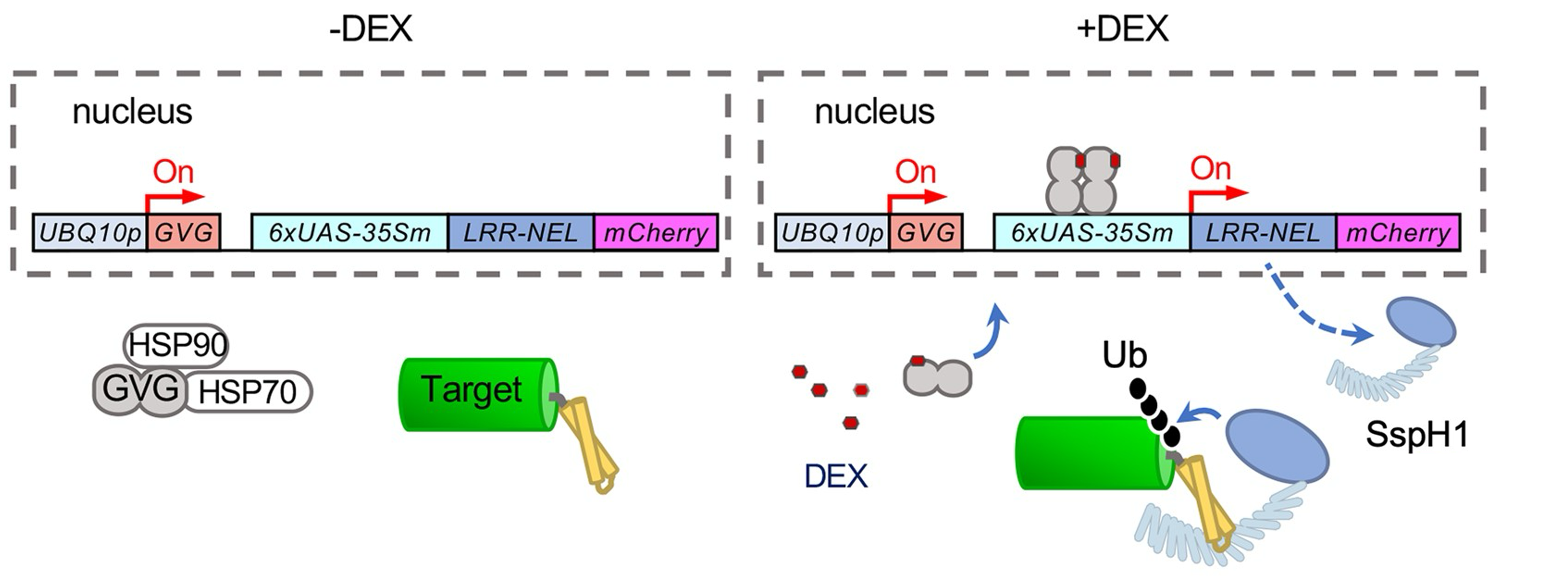 Targeting a given protein for degradation at a specific time can be very useful when investigating its function in the cellular context. In this paper, Huang et al. present an inducible protein degradation system tailored for plants, named E3-DART. The system is based on the specific interaction between the type III bacterial effector Salmonella-specific protein H1 (SspH1) and the homology region 1b (HR1b) of its human target, the protein kinase N1 (PKN1). SspH1 binds HR1b with high specificity and ubiquintinates it, thus targeting it for degradation. The authors leveraged this interaction to engineer a minimal version of SspH1 capable of targeting any HR1b-tagged protein, both in transient expression assays (agroinfiltration) and in stably trasnformed lines. To make the system inducible, they used the GAL4-VP16-GVG transcriptional module, where the GVG transcription factor, under the control of a ubiquitin promoter, is activated by the application of dexamethanose and thereafter binds to the GAL4 promoter to induce the transcription of their engineered SspH1 protein. In turn, SspH1 ubiquitinates any protein containing the HR1b domain and depletes its amount in as short as 3 hours. E3-DART is to date one of the only two protein degradation systems developed for plants and holds great potential for the conditional knock-out of desired proteins in a short timeframe. (Summary by Carlo Pasini @Crl_Psn) Plant Cell 10.1093/plcell/koae072
Targeting a given protein for degradation at a specific time can be very useful when investigating its function in the cellular context. In this paper, Huang et al. present an inducible protein degradation system tailored for plants, named E3-DART. The system is based on the specific interaction between the type III bacterial effector Salmonella-specific protein H1 (SspH1) and the homology region 1b (HR1b) of its human target, the protein kinase N1 (PKN1). SspH1 binds HR1b with high specificity and ubiquintinates it, thus targeting it for degradation. The authors leveraged this interaction to engineer a minimal version of SspH1 capable of targeting any HR1b-tagged protein, both in transient expression assays (agroinfiltration) and in stably trasnformed lines. To make the system inducible, they used the GAL4-VP16-GVG transcriptional module, where the GVG transcription factor, under the control of a ubiquitin promoter, is activated by the application of dexamethanose and thereafter binds to the GAL4 promoter to induce the transcription of their engineered SspH1 protein. In turn, SspH1 ubiquitinates any protein containing the HR1b domain and depletes its amount in as short as 3 hours. E3-DART is to date one of the only two protein degradation systems developed for plants and holds great potential for the conditional knock-out of desired proteins in a short timeframe. (Summary by Carlo Pasini @Crl_Psn) Plant Cell 10.1093/plcell/koae072