Plant Science Research Weekly: March 27
Review. Signalling pathways underlying nitrogen-dependent changes in root system architecture: from model to crop species
 Nitrogen (N) is one of the seventeen essential nutrients for a plant to complete its life cycle and is one of the most important determinants of productivity of various crops globally. Nitrate (NO3‑) and ammonium (NH4+) are the major plant-available forms of N. The spatiotemporal heterogeneity of N is tackled by the plastic responses of the plant root system which helps the plant explore for N. Jia and von Wirén have reviewed the signalling pathways underlying the modulation in root system architecture due to internal and external N signals. First the authors discuss the plant root system’s response to availability of N in the soil. When a root comes across a localized N-rich zone, NO3‑ or NH4+ acts as a signalling molecule and stimulates lateral root elongation; branching is stimulated through the transceptor NRT1.1/NFP6.3 or AMT transporters, respectively which is termed as foraging. When a root comes across a homogenous availability of N, the response of the root growth is dose-dependent; a moderate availability promotes the root growth where as a high availability suppresses the same. The authors also discuss the mechanisms underlying root system’s response during N scarcity. Plants stimulate root growth when the scarcity is mild; on the other hand, when the scarcity is severe, plants adopt an economical strategy of survival by halting their root growth and developement. Understanding the underlying genetic factors and mechanisms is essential for enhancing the nutrient use efficiency in economically important crops through various crop improvement programmes using breeding and genome editing. (Summary by Hari Gowthem G) J. Exp. Bot. 10.1093/jxb/eraa033
Nitrogen (N) is one of the seventeen essential nutrients for a plant to complete its life cycle and is one of the most important determinants of productivity of various crops globally. Nitrate (NO3‑) and ammonium (NH4+) are the major plant-available forms of N. The spatiotemporal heterogeneity of N is tackled by the plastic responses of the plant root system which helps the plant explore for N. Jia and von Wirén have reviewed the signalling pathways underlying the modulation in root system architecture due to internal and external N signals. First the authors discuss the plant root system’s response to availability of N in the soil. When a root comes across a localized N-rich zone, NO3‑ or NH4+ acts as a signalling molecule and stimulates lateral root elongation; branching is stimulated through the transceptor NRT1.1/NFP6.3 or AMT transporters, respectively which is termed as foraging. When a root comes across a homogenous availability of N, the response of the root growth is dose-dependent; a moderate availability promotes the root growth where as a high availability suppresses the same. The authors also discuss the mechanisms underlying root system’s response during N scarcity. Plants stimulate root growth when the scarcity is mild; on the other hand, when the scarcity is severe, plants adopt an economical strategy of survival by halting their root growth and developement. Understanding the underlying genetic factors and mechanisms is essential for enhancing the nutrient use efficiency in economically important crops through various crop improvement programmes using breeding and genome editing. (Summary by Hari Gowthem G) J. Exp. Bot. 10.1093/jxb/eraa033
Hornwort genomes
 A major update in plant genome information is taking place. Two independent groups have published genomes of hornwort species from the Anthoceros genus. Both papers arrive at similar conclusions supporting the model of a single “Setaphyta” clade, with hornworts sister to liverworts and mosses. These genomes shed light on the conserved repertoire of bryophyte genes and help to elucidate the genetic features of the most recent common ancestor of embryophytes (land plants). Anthoceros genomes lack whole-genome duplications as do most liverworts and, like Marchantia polymorpha, present low genetic redundancy. In the Li et al. paper, three genomes from two different Anthoceros species were sequenced. Additionally, they provide a chromosome-scale assembly of the A. agresti genome, revealing an even distribution of genes and repetitive regions without enrichment in the centromeric regions, an unusual feature that seems to be similar to other bryophyte genomes. They also identified genes associated with cyanobacteria symbiosis, and discuss possible genes involved in pyrenoid carbon-concentration mechanism. The Zhang et al. paper presents the genome of a third species, A. angustus. They highlight horizontal gene transfer events that could be relevant for metabolic processes. Without doubt, Anthoceros genomes will be an ineludible reference for evolutionary studies aiming to understand plant macroevolution. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-020-0618-2 and 10.1038/s41477-019-0588-4
A major update in plant genome information is taking place. Two independent groups have published genomes of hornwort species from the Anthoceros genus. Both papers arrive at similar conclusions supporting the model of a single “Setaphyta” clade, with hornworts sister to liverworts and mosses. These genomes shed light on the conserved repertoire of bryophyte genes and help to elucidate the genetic features of the most recent common ancestor of embryophytes (land plants). Anthoceros genomes lack whole-genome duplications as do most liverworts and, like Marchantia polymorpha, present low genetic redundancy. In the Li et al. paper, three genomes from two different Anthoceros species were sequenced. Additionally, they provide a chromosome-scale assembly of the A. agresti genome, revealing an even distribution of genes and repetitive regions without enrichment in the centromeric regions, an unusual feature that seems to be similar to other bryophyte genomes. They also identified genes associated with cyanobacteria symbiosis, and discuss possible genes involved in pyrenoid carbon-concentration mechanism. The Zhang et al. paper presents the genome of a third species, A. angustus. They highlight horizontal gene transfer events that could be relevant for metabolic processes. Without doubt, Anthoceros genomes will be an ineludible reference for evolutionary studies aiming to understand plant macroevolution. (Summary by Facundo Romani) Nature Plants 10.1038/s41477-020-0618-2 and 10.1038/s41477-019-0588-4
Mars1 kinase signaling in the chloroplast unfolded protein response
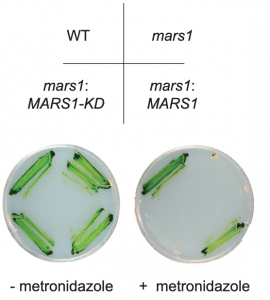 In stressful situations, such as high light and nutrient scarcity, the chloroplast may experience increased proteotoxicity due to a surge in damaging reactive oxygen species. In response, a signal is sent to the nucleus to increase production of many proteins, including proteases and chaperones to help mitigate the damage. This damage control pathway has been termed the chloroplast unfolded protein response (cpUPR) and is thought to play a key role in regulating plastid protein homeostasis, but the molecular basis for cpUPR signaling has been largely unknown. To understand how the cpUPR turns on, Perlaza et al. set up a genetic screen in the green alga Chlamydomonas based on conditional cpUPR activation combined with fluorescent read-out of a nuclear-encoded reporter gene. Through this screen they identified Mars1, a kinase specifically required for communication between the chloroplast and nucleus in response to proteotoxic stress. Break-down of plastid-to-nucleus communication in the mars1 mutant caused a failure to activate the cpUPR transcriptional response and severely reduced growth under high light intensity. Conversely, Mars1 overexpression protected cells against normally growth-inhibitory conditions, suggesting that cpUPR engineering could be a viable strategy to increase plant stress tolerance. (Summary by Frej Tulin) eLIFE 10.7554/eLife.49577
In stressful situations, such as high light and nutrient scarcity, the chloroplast may experience increased proteotoxicity due to a surge in damaging reactive oxygen species. In response, a signal is sent to the nucleus to increase production of many proteins, including proteases and chaperones to help mitigate the damage. This damage control pathway has been termed the chloroplast unfolded protein response (cpUPR) and is thought to play a key role in regulating plastid protein homeostasis, but the molecular basis for cpUPR signaling has been largely unknown. To understand how the cpUPR turns on, Perlaza et al. set up a genetic screen in the green alga Chlamydomonas based on conditional cpUPR activation combined with fluorescent read-out of a nuclear-encoded reporter gene. Through this screen they identified Mars1, a kinase specifically required for communication between the chloroplast and nucleus in response to proteotoxic stress. Break-down of plastid-to-nucleus communication in the mars1 mutant caused a failure to activate the cpUPR transcriptional response and severely reduced growth under high light intensity. Conversely, Mars1 overexpression protected cells against normally growth-inhibitory conditions, suggesting that cpUPR engineering could be a viable strategy to increase plant stress tolerance. (Summary by Frej Tulin) eLIFE 10.7554/eLife.49577
Roles for CHROMATIN REMODELING 4 in Arabidopsis floral transition
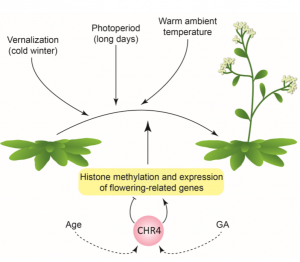 The time at which flowers appear is critical for plant reproductive success. As such, the vegetative to reproductive growth transition is governed by several cues: environmental (photoperiod, temperature) and endogenous (gibberellins, age). Here, Sang et al. used an elegant forward-genetics approach to identify uncharacterized regulators of the Arabidopsis endogenous flowering time pathways. A quintuple mutant (svp-41 flc-3 ft-10 tsf-1 soc1-2) was formed to ensure that floral initiation could only occur endogenously. In other words, the key regulators that induce flowering under high temperatures and long-day conditions were mutated. This quintuple mutant was further mutagenized and lines displaying late-flowering phenotypes were investigated. In this manner, CHROMATIN REMODELING 4 (CHR4) was identified as a positive regulator of floral transition. Immunoprecipitation experiments suggest that CHR4 functions in multimeric complexes with known regulators of floral meristem identity and transition such as the MADS, SPL and AP2 transcription factor families. Furthermore, in chr4 plants, ChIP-seq revealed several genomic loci that possess altered histone methylation patterns and RNA-seq showed that expression levels of various floral regulators are modified. Future studies will endeavor to determine the CHR4 mechanism of regulating gene expression. (Summary by Caroline Dowling) Plant Cell 10.1105/tpc.19.00992 (Image: Hanna Hõrak, The Plant Cell In Brief 10.1105/tpc.20.00196)
The time at which flowers appear is critical for plant reproductive success. As such, the vegetative to reproductive growth transition is governed by several cues: environmental (photoperiod, temperature) and endogenous (gibberellins, age). Here, Sang et al. used an elegant forward-genetics approach to identify uncharacterized regulators of the Arabidopsis endogenous flowering time pathways. A quintuple mutant (svp-41 flc-3 ft-10 tsf-1 soc1-2) was formed to ensure that floral initiation could only occur endogenously. In other words, the key regulators that induce flowering under high temperatures and long-day conditions were mutated. This quintuple mutant was further mutagenized and lines displaying late-flowering phenotypes were investigated. In this manner, CHROMATIN REMODELING 4 (CHR4) was identified as a positive regulator of floral transition. Immunoprecipitation experiments suggest that CHR4 functions in multimeric complexes with known regulators of floral meristem identity and transition such as the MADS, SPL and AP2 transcription factor families. Furthermore, in chr4 plants, ChIP-seq revealed several genomic loci that possess altered histone methylation patterns and RNA-seq showed that expression levels of various floral regulators are modified. Future studies will endeavor to determine the CHR4 mechanism of regulating gene expression. (Summary by Caroline Dowling) Plant Cell 10.1105/tpc.19.00992 (Image: Hanna Hõrak, The Plant Cell In Brief 10.1105/tpc.20.00196)
A feedforward loop controls vascular regeneration and tissue repair through local auxin biosynthesis
 Plant cells are entrapped in rigid cell walls, so morphogenesis relies on asymmetric cell division (ACD) and positional cues to regulate tissue patterning. The Arabidopsis phloem is a good system to study tissue patterning due to its relatively simple composition: sieve elements (SEs) and companion cells (CCs) lineages. Although the regulatory programs that control phloem development are well known, the molecular mechanism that coordinates the ACDs that give rise to the SEs and CCs lineages is obscure. In this study, Kim et al. reveal how the SHORTROOT (SHR) mobile transcription factor orchestrates the development of SEs and CCs precursors in two different spatial contexts. By analyzing stele expression data and inducible lines the authors determined that NAC-REGULATED SEED MORPHOLOGY 1 (NARS1) and SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 (SND2) are direct targets of SHR. Subsequent analyses showed that SHR movement into the endodermis, where it interacts with SCARECROW (SCR), indirectly activates transcription of NARS1 through microRNA165/166 to control procambial cell proliferation, and thus the ACD required for CCs. Movement of SHR into the phloem however activates transcription of NARS1 in the root differentiation zone, which, acting as a top-down mobile signal, directly activates SND2 expression in the root meristem to further amplify NARS1 expression, forming a positive feedforward loop to regulate ACD of SEs precursors. This regulatory program sheds light on the complex molecular mechanisms required for tissue patterning. (Summary by Jesus Leon) Plant Cell 10.1105/tpc.19.00455
Plant cells are entrapped in rigid cell walls, so morphogenesis relies on asymmetric cell division (ACD) and positional cues to regulate tissue patterning. The Arabidopsis phloem is a good system to study tissue patterning due to its relatively simple composition: sieve elements (SEs) and companion cells (CCs) lineages. Although the regulatory programs that control phloem development are well known, the molecular mechanism that coordinates the ACDs that give rise to the SEs and CCs lineages is obscure. In this study, Kim et al. reveal how the SHORTROOT (SHR) mobile transcription factor orchestrates the development of SEs and CCs precursors in two different spatial contexts. By analyzing stele expression data and inducible lines the authors determined that NAC-REGULATED SEED MORPHOLOGY 1 (NARS1) and SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 2 (SND2) are direct targets of SHR. Subsequent analyses showed that SHR movement into the endodermis, where it interacts with SCARECROW (SCR), indirectly activates transcription of NARS1 through microRNA165/166 to control procambial cell proliferation, and thus the ACD required for CCs. Movement of SHR into the phloem however activates transcription of NARS1 in the root differentiation zone, which, acting as a top-down mobile signal, directly activates SND2 expression in the root meristem to further amplify NARS1 expression, forming a positive feedforward loop to regulate ACD of SEs precursors. This regulatory program sheds light on the complex molecular mechanisms required for tissue patterning. (Summary by Jesus Leon) Plant Cell 10.1105/tpc.19.00455
The embryo sheath is an anti-adhesive structure that facilitates cotyledon emergence during germination in Arabidopsis ($)
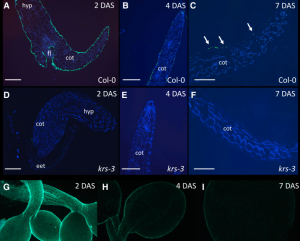 The embryo sheath is a glycoprotein structure formed during seed maturation in Arabidopsis thaliana. However, its function and its persistence after germination have been uncertain. Here, Doll et al. examined germination and seedling establishment of krs mutants deficient in the KERBEROS peptide that had been previously identified as contributing to embryo sheath formation. By using immunolabelling, the authors demonstrate that the embryo sheath is maintained on the seedling surface after germination. However, in the krs seedlings this structure is missing. Moreover, Doll et al. found that the genetic basis of sheath and cuticle formation is independent, given the cuticle-deficient mutants do have the embryo sheath. Interestingly, the authors found the embryo sheath facilitated cotyledon emergence because it reduces embryo adhesiveness with the rest of seed internal components. Moreover, competition assays showed that wild type seedlings outperformed the krs mutants, suggesting that the embryo sheath has a significant impact on seedling establishment. (Summary by Carlos A. Ordóñez-Parra) Curr. Biol. j.cub.2019.12.057
The embryo sheath is a glycoprotein structure formed during seed maturation in Arabidopsis thaliana. However, its function and its persistence after germination have been uncertain. Here, Doll et al. examined germination and seedling establishment of krs mutants deficient in the KERBEROS peptide that had been previously identified as contributing to embryo sheath formation. By using immunolabelling, the authors demonstrate that the embryo sheath is maintained on the seedling surface after germination. However, in the krs seedlings this structure is missing. Moreover, Doll et al. found that the genetic basis of sheath and cuticle formation is independent, given the cuticle-deficient mutants do have the embryo sheath. Interestingly, the authors found the embryo sheath facilitated cotyledon emergence because it reduces embryo adhesiveness with the rest of seed internal components. Moreover, competition assays showed that wild type seedlings outperformed the krs mutants, suggesting that the embryo sheath has a significant impact on seedling establishment. (Summary by Carlos A. Ordóñez-Parra) Curr. Biol. j.cub.2019.12.057
“The secret is how to die!”: CPK1 controls cell death via phosphorylation of ORE1
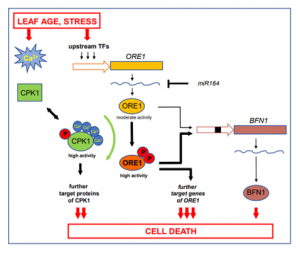 Cell death, like all other cellular processes, is controlled by a highly complex signaling network. A search for the targets of Arabidopsis calcium dependent protein kinase1 (CPK1) by Durian and co-workers has revealed that it phosphorylates ORESARA1 (ORE1), a master regulator of leaf senescence responses, in a region necessary for its downstream gene activation. Characterization of this CPK1-ORE1 interaction showed that CPK1 promotes cell death through ORE1 by positively activating it, and thereby its target genes. This process is independent of calcium ion (Ca2+) concentration as the authors used a constitutively active, Ca2+-independent variant of CPK1 (named CPK1-VK), with only its N-terminal and kinase domains. This report presents a new regulatory network for ORE1 and calls into question the requirement of Ca2+ for CPK1 activity. (Summary by Pavithran Narayanan) Plant Cell 10.1105/tpc.19.00810. (Quote from Dan Brown’s ‘The Lost Symbol’).
Cell death, like all other cellular processes, is controlled by a highly complex signaling network. A search for the targets of Arabidopsis calcium dependent protein kinase1 (CPK1) by Durian and co-workers has revealed that it phosphorylates ORESARA1 (ORE1), a master regulator of leaf senescence responses, in a region necessary for its downstream gene activation. Characterization of this CPK1-ORE1 interaction showed that CPK1 promotes cell death through ORE1 by positively activating it, and thereby its target genes. This process is independent of calcium ion (Ca2+) concentration as the authors used a constitutively active, Ca2+-independent variant of CPK1 (named CPK1-VK), with only its N-terminal and kinase domains. This report presents a new regulatory network for ORE1 and calls into question the requirement of Ca2+ for CPK1 activity. (Summary by Pavithran Narayanan) Plant Cell 10.1105/tpc.19.00810. (Quote from Dan Brown’s ‘The Lost Symbol’).
The cis-regulatory codes of response to combined heat and drought stress
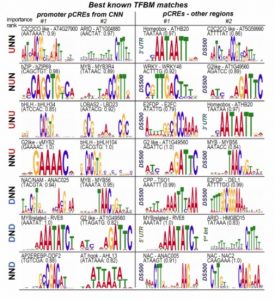 As sessile organisms, plants must not only respond to a single stress, but multiple stresses at the same time. To understand the DNA regulatory elements that mediate the transcriptional response to heat, drought and combined heat and drought stress, Azodi et al. utilized the known transcription factor binding motifs (TFBM) and enrichment based putative cis-regulatory elements (pCREs) to define the cis-regulatory codes. The authors found that the genes responsive to combined heat and drought stress have unique regulatory codes that are needed to fine tune the transcriptional response. By using a deep learning algorithm, convolutional neural networks (CNNs), the authors could predict and improve the patterns of response to single and combined stress. They also summarize that combining pCREs identified outside of the promoter region are better predictive response to single/combined stresses. In conclusion, they highlight the complexity of the identified cis-regulatory codes to characterize some of the most important pCREs in response to single/combined stress. The approaches used in this study could be used to understand the regulatory network of developmental and stress induced responses in other organisms. (Summary by Min May Wong) bioRxiv 10.1101/2020.02.28.969261
As sessile organisms, plants must not only respond to a single stress, but multiple stresses at the same time. To understand the DNA regulatory elements that mediate the transcriptional response to heat, drought and combined heat and drought stress, Azodi et al. utilized the known transcription factor binding motifs (TFBM) and enrichment based putative cis-regulatory elements (pCREs) to define the cis-regulatory codes. The authors found that the genes responsive to combined heat and drought stress have unique regulatory codes that are needed to fine tune the transcriptional response. By using a deep learning algorithm, convolutional neural networks (CNNs), the authors could predict and improve the patterns of response to single and combined stress. They also summarize that combining pCREs identified outside of the promoter region are better predictive response to single/combined stresses. In conclusion, they highlight the complexity of the identified cis-regulatory codes to characterize some of the most important pCREs in response to single/combined stress. The approaches used in this study could be used to understand the regulatory network of developmental and stress induced responses in other organisms. (Summary by Min May Wong) bioRxiv 10.1101/2020.02.28.969261
A plant kinase exploited by a bacterial effector
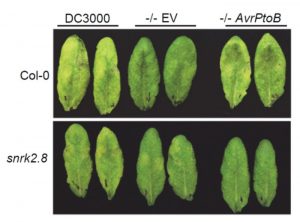 Pathogens have evolved a suite of effector proteins that are secreted into plants and aid successful colonization. AvrPtoB is a well-conserved effector of pathogenic Pseudomonas syringae strains that targets multiple plant defense-related proteins and is required for pathogen virulence and bacterial growth in plants. Phosphorylation of AvrPtoB in plants is important for its virulence functions, but the mechanism by which the effector is phosphorylated is unknown. Lei et al. demonstrated that the plant kinase SnRK2.8, a member of a widely conserved plant kinase family, interacts with AvrPtoB and phosphorylates this effector protein in Arabidopsis thaliana. The virulence functions of AvrPtoB were abolished in the absence of SnRK2.8 or when the phosphorylation sites are blocked. SnRK2.8 was proposed to phosphorylate the key plant defense regulator NPR1 to mediate plant immunity. This mechanism appears to be exploited by AvrPtoB, which, upon phosphorylation, degrades NPR1. This study identifies the first plant kinase exploited by a bacterial pathogen to phosphorylate and activate an effector protein illuminating an evolutionary arms race between plants and pathogens. (Summary by Tatsuya Nobori) bioRxiv 10.1101/2020.03.21.001826v1
Pathogens have evolved a suite of effector proteins that are secreted into plants and aid successful colonization. AvrPtoB is a well-conserved effector of pathogenic Pseudomonas syringae strains that targets multiple plant defense-related proteins and is required for pathogen virulence and bacterial growth in plants. Phosphorylation of AvrPtoB in plants is important for its virulence functions, but the mechanism by which the effector is phosphorylated is unknown. Lei et al. demonstrated that the plant kinase SnRK2.8, a member of a widely conserved plant kinase family, interacts with AvrPtoB and phosphorylates this effector protein in Arabidopsis thaliana. The virulence functions of AvrPtoB were abolished in the absence of SnRK2.8 or when the phosphorylation sites are blocked. SnRK2.8 was proposed to phosphorylate the key plant defense regulator NPR1 to mediate plant immunity. This mechanism appears to be exploited by AvrPtoB, which, upon phosphorylation, degrades NPR1. This study identifies the first plant kinase exploited by a bacterial pathogen to phosphorylate and activate an effector protein illuminating an evolutionary arms race between plants and pathogens. (Summary by Tatsuya Nobori) bioRxiv 10.1101/2020.03.21.001826v1
How Marchantia polymorpha avoids bug bites
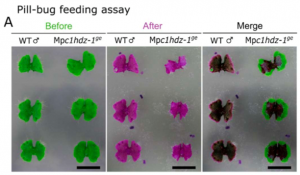 Plants took hundreds of million years to evolve from aquatic to land environments. Biotic and abiotic stress adaptation contributed to the transition. In this preprint, Romani et al. elucidated functions of the transcription factor CLASS I HOMEODOMAIN LEUCINE-ZIPPER (C1HDZ) in the early land plant Marchantia polymorpha. Despite having all the conserved protein motifs of other members of this family, MpC1HDZ does not seem to contribute to abiotic stress tolerance whereas C1HDZ genes in flowering plants are known to regulate abiotic stress responses. Interestingly, knocking out MpC1HDZ greatly decreased oil body numbers, and transcriptome analyses of the mutants showed that majority of the down-regulated genes were associated with secondary metabolism. Moreover, the mutants with early stop codons (e.g. Mpc1hdz-1ge) were preferred by starved pill bugs, a terrestrial isopod, and thallus extract of the mutants showed decreased antibiotic activity against Bacillus subtilis. This study shows genetic evidence in support of the century-old hypothesis that liverwort oil bodies function in defense against arthropod herbivores. Lastly, the authors propose an oil body cell differentiation model that is mediated by MpC1HDZ expression. (Summary by Yun-Ting Kao) bioRxiv 10.1101/2020.03.02.971010
Plants took hundreds of million years to evolve from aquatic to land environments. Biotic and abiotic stress adaptation contributed to the transition. In this preprint, Romani et al. elucidated functions of the transcription factor CLASS I HOMEODOMAIN LEUCINE-ZIPPER (C1HDZ) in the early land plant Marchantia polymorpha. Despite having all the conserved protein motifs of other members of this family, MpC1HDZ does not seem to contribute to abiotic stress tolerance whereas C1HDZ genes in flowering plants are known to regulate abiotic stress responses. Interestingly, knocking out MpC1HDZ greatly decreased oil body numbers, and transcriptome analyses of the mutants showed that majority of the down-regulated genes were associated with secondary metabolism. Moreover, the mutants with early stop codons (e.g. Mpc1hdz-1ge) were preferred by starved pill bugs, a terrestrial isopod, and thallus extract of the mutants showed decreased antibiotic activity against Bacillus subtilis. This study shows genetic evidence in support of the century-old hypothesis that liverwort oil bodies function in defense against arthropod herbivores. Lastly, the authors propose an oil body cell differentiation model that is mediated by MpC1HDZ expression. (Summary by Yun-Ting Kao) bioRxiv 10.1101/2020.03.02.971010
Evidence for physiological seed dormancy cycling in the woody shrub Asterolasia buxifolia and its ecological significance in fire‐prone systems ($)
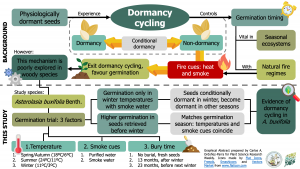 Physiologically dormant seeds shift between dormancy (i.e., unable to germinate), conditional dormancy (i.e., germination restricted to a narrow set of conditions), and non-dormancy (i.e., germination under a wide range of conditions) in response to environmental changes. This mechanism –known as dormancy cycling– ensures seeds are non-dormant under favorable germination conditions but become dormant when favorable conditions resume. In fire-prone ecosystems, heat and smoke guarantee seeds exit dormancy cycling and germinate. However, this phenomenon is seldom investigated in woody species. In this study, Collette and Ooi found Asterolasia buxifolia Benth. experiences dormancy cycling. The authors carried out an experiment using fresh seeds and seeds buried and recovered after the next winter, and before the following one. The experiment assessed germination in three temperatures (spring/autumn, summer, and winter) and the effect of smoke cues (distilled water vs. smoke water). Seeds only germinated at winter temperatures with smoke water. Also, germination was higher in seeds recovered before winter than those recovered after winter. These results indicate that the seeds of A. buxifolia have an annual dormancy cycle ensuring winter germination, after natural fires cues are expected to occur. (Summary by Carlos A. Ordóñez-Parra) Plant Biol. 10.1111/plb.13105
Physiologically dormant seeds shift between dormancy (i.e., unable to germinate), conditional dormancy (i.e., germination restricted to a narrow set of conditions), and non-dormancy (i.e., germination under a wide range of conditions) in response to environmental changes. This mechanism –known as dormancy cycling– ensures seeds are non-dormant under favorable germination conditions but become dormant when favorable conditions resume. In fire-prone ecosystems, heat and smoke guarantee seeds exit dormancy cycling and germinate. However, this phenomenon is seldom investigated in woody species. In this study, Collette and Ooi found Asterolasia buxifolia Benth. experiences dormancy cycling. The authors carried out an experiment using fresh seeds and seeds buried and recovered after the next winter, and before the following one. The experiment assessed germination in three temperatures (spring/autumn, summer, and winter) and the effect of smoke cues (distilled water vs. smoke water). Seeds only germinated at winter temperatures with smoke water. Also, germination was higher in seeds recovered before winter than those recovered after winter. These results indicate that the seeds of A. buxifolia have an annual dormancy cycle ensuring winter germination, after natural fires cues are expected to occur. (Summary by Carlos A. Ordóñez-Parra) Plant Biol. 10.1111/plb.13105