Plant Science Research Weekly: March 1, 2024
Review: SynBio takes on roots and the rhizosphere
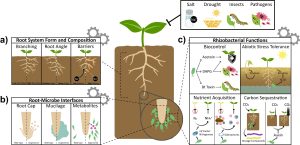 This is an excellent introduction to how synthetic biology can be used to program plants for climate resilience by engineering them to respond predictably and in ways beyond those that evolution has explored, through the use of controllable synthetic gene circuits. Ragland et al. describe how precise and specific transcription factors can target synthetic promoter elements with little bleed-through to other genes, providing very fine and predictable control of gene expression and eliminating confounding pleiotropic effects. Combining these elements as Boolean logic gates can produce expression patterns in cells where conditions are absent (NOR gates) or NIMPLY gates where expression occurs where one circuit is on and the other off. After describing some of these synthetic gene circuits, the article discusses how they can be used to modify root system architecture, anatomy, and nutrient uptake. Several key genes have been identified with large impacts on root development, such as lateral root initiation or gravity setpoint angle, which in turn affect water and nutrient uptake. Genes that affect nutrient uptake, soil penetrance, and exudation of molecules into the soil are also promising candidates for synthetic biology-based engineering. Finally, plant productivity can be promoted by the potentially quicker engineering of soil microbes, with examples discussed that can impact plant biotic and abiotic interactions, as well as nutrient acquisition. Through the use of the design-build-test-learn cycle, its likely that we’ll be seeing rapid progress in the engineering of plant root systems. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-024-45272-5
This is an excellent introduction to how synthetic biology can be used to program plants for climate resilience by engineering them to respond predictably and in ways beyond those that evolution has explored, through the use of controllable synthetic gene circuits. Ragland et al. describe how precise and specific transcription factors can target synthetic promoter elements with little bleed-through to other genes, providing very fine and predictable control of gene expression and eliminating confounding pleiotropic effects. Combining these elements as Boolean logic gates can produce expression patterns in cells where conditions are absent (NOR gates) or NIMPLY gates where expression occurs where one circuit is on and the other off. After describing some of these synthetic gene circuits, the article discusses how they can be used to modify root system architecture, anatomy, and nutrient uptake. Several key genes have been identified with large impacts on root development, such as lateral root initiation or gravity setpoint angle, which in turn affect water and nutrient uptake. Genes that affect nutrient uptake, soil penetrance, and exudation of molecules into the soil are also promising candidates for synthetic biology-based engineering. Finally, plant productivity can be promoted by the potentially quicker engineering of soil microbes, with examples discussed that can impact plant biotic and abiotic interactions, as well as nutrient acquisition. Through the use of the design-build-test-learn cycle, its likely that we’ll be seeing rapid progress in the engineering of plant root systems. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-024-45272-5
Review: Long noncoding RNAs and the art of being influential without protein
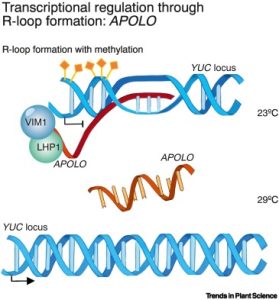 Advances in sequencing technologies have led to a tremendous growth in the number of long noncoding RNAs (lncRNAs) being identified in plants, but identifying their function lags behind. Here, Ramirez Gonzales and Blom et al. draw on lessons from studies in mammalian systems and highlight several known functions of lncRNAs. One of the more well-known plant lncRNA is COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), a component of the vernalization response that affects histone methylation and transcriptional silencing at FLOWERING LOCUS C (FLC). Other lncRNAs in plants and mammals also have epigenetic effects including chromatin methylation. As nucleic acids, lncRNAs can directly interact with DNA and RNA including microRNAs (miRNAs) through base pairing. One of the better-known examples of this involves the interaction between the lncRNA INDUCED BY PHOSPHATE STARVATION1 (IPS1) and miR399. This interaction prevents miR399 from binding to its “real” target, the mRNA PHO2, thus regulating phosphate uptake. Other lncRNA functions that are discussed include splicing, other forms of transcriptional regulation, and acting as a mobile signal. Besides being a really clear and thorough overview of lncRNAs, this review highlights potential applications of lncRNAs in plants. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.01.006
Advances in sequencing technologies have led to a tremendous growth in the number of long noncoding RNAs (lncRNAs) being identified in plants, but identifying their function lags behind. Here, Ramirez Gonzales and Blom et al. draw on lessons from studies in mammalian systems and highlight several known functions of lncRNAs. One of the more well-known plant lncRNA is COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), a component of the vernalization response that affects histone methylation and transcriptional silencing at FLOWERING LOCUS C (FLC). Other lncRNAs in plants and mammals also have epigenetic effects including chromatin methylation. As nucleic acids, lncRNAs can directly interact with DNA and RNA including microRNAs (miRNAs) through base pairing. One of the better-known examples of this involves the interaction between the lncRNA INDUCED BY PHOSPHATE STARVATION1 (IPS1) and miR399. This interaction prevents miR399 from binding to its “real” target, the mRNA PHO2, thus regulating phosphate uptake. Other lncRNA functions that are discussed include splicing, other forms of transcriptional regulation, and acting as a mobile signal. Besides being a really clear and thorough overview of lncRNAs, this review highlights potential applications of lncRNAs in plants. (Summary by Mary Williams @PlantTeaching) Trends Plant Sci. 10.1016/j.tplants.2024.01.006
Review: Development and evolution of the Asteraceae inflorescence
 Asteraceae, also known as Compositae or the daisy family, is one of the largest plant families and comprises 10% of all flowering plants. Members of this family are found in habitats worldwide. The unique inflorescence, called a capitulum, is a key innovation of the family and contributes to its success. This highly compressed head-like structure can consist of a single flower but usually is composed of multiple florets. In this new review, Zhang and Elomaa delve into the development and evolution of the capitulum, shedding light on molecular and genetic studies that have expanded our understanding of its formation such as exploring the patterning and maturation of florets within the capitulum, including the iconic phyllotactic arrangement of florets in Fibonacci numbers of spirals. While much has been uncovered about the capitulum’s evolution and organization, there is still a large numberr of species to explore. The paper emphasizes Asteraceae’s potential as a model for evolutionary developmental studies, utilizing its diverse morphology and genetic networks to advance crop breeding efforts. (Summary by Villő Bernád) New Phytol. 10.1111/nph.19590
Asteraceae, also known as Compositae or the daisy family, is one of the largest plant families and comprises 10% of all flowering plants. Members of this family are found in habitats worldwide. The unique inflorescence, called a capitulum, is a key innovation of the family and contributes to its success. This highly compressed head-like structure can consist of a single flower but usually is composed of multiple florets. In this new review, Zhang and Elomaa delve into the development and evolution of the capitulum, shedding light on molecular and genetic studies that have expanded our understanding of its formation such as exploring the patterning and maturation of florets within the capitulum, including the iconic phyllotactic arrangement of florets in Fibonacci numbers of spirals. While much has been uncovered about the capitulum’s evolution and organization, there is still a large numberr of species to explore. The paper emphasizes Asteraceae’s potential as a model for evolutionary developmental studies, utilizing its diverse morphology and genetic networks to advance crop breeding efforts. (Summary by Villő Bernád) New Phytol. 10.1111/nph.19590
Moss phenotype unaffected by removal of repetitive sequences from genome
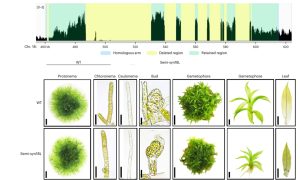 Genome complexity in multicellular organisms is often associated with repetitive sequences from Transposable Elements (TEs). TE function, importance for genome integrity, and dispensability have not been completely characterized. In prokaryotes and simple eukaryotes, genome synthesis (production of synthetic chromosomes and genomes) has proven to be a useful tool to understand genome organisation and function. As a prelude to the development of synthetic genomes in a multicellular organism. Physcomitrium (Physcomitrella) patens is a well-established multicellular model organism that is more complex than unicellular organisms due to greater genome size, gene number and epigenetic diversity. Chen and collaborators investigated the effect of decreased repetitive sequences by eliminating 55.8% of a 155 kb region of chromosome 18 in Physcomitrella. They used various methods to reassemble the DNA, including homologous recombination which was highly effective. Surprisingly, the phenotype of the transformed plant was comparable to the wild type (WT) even when growing under salt and osmotic stress. There was not much difference in histone marks related to active transcription between the synthetic and WT genomic regions, however marks associated with TEs abundance (H3K9me2) were almost completely removed, probably due to the lack of repetitive elements. On the other hand, chromatin interaction did show changes in the redesigned and neighbouring regions, demonstrating that genome redesign can alter chromosome conformation. These studies show that repetitive sequences can be removed without affecting morphology, development, and stress resistance, paving the way for the Synthetic Moss (SynMoss) project. (Summary by Rigel Salinas-Gamboa @Rigelitactica). Nature Plants 10.1038/s41477-023-01595-7
Genome complexity in multicellular organisms is often associated with repetitive sequences from Transposable Elements (TEs). TE function, importance for genome integrity, and dispensability have not been completely characterized. In prokaryotes and simple eukaryotes, genome synthesis (production of synthetic chromosomes and genomes) has proven to be a useful tool to understand genome organisation and function. As a prelude to the development of synthetic genomes in a multicellular organism. Physcomitrium (Physcomitrella) patens is a well-established multicellular model organism that is more complex than unicellular organisms due to greater genome size, gene number and epigenetic diversity. Chen and collaborators investigated the effect of decreased repetitive sequences by eliminating 55.8% of a 155 kb region of chromosome 18 in Physcomitrella. They used various methods to reassemble the DNA, including homologous recombination which was highly effective. Surprisingly, the phenotype of the transformed plant was comparable to the wild type (WT) even when growing under salt and osmotic stress. There was not much difference in histone marks related to active transcription between the synthetic and WT genomic regions, however marks associated with TEs abundance (H3K9me2) were almost completely removed, probably due to the lack of repetitive elements. On the other hand, chromatin interaction did show changes in the redesigned and neighbouring regions, demonstrating that genome redesign can alter chromosome conformation. These studies show that repetitive sequences can be removed without affecting morphology, development, and stress resistance, paving the way for the Synthetic Moss (SynMoss) project. (Summary by Rigel Salinas-Gamboa @Rigelitactica). Nature Plants 10.1038/s41477-023-01595-7
The Marchantia transcription factor atlas
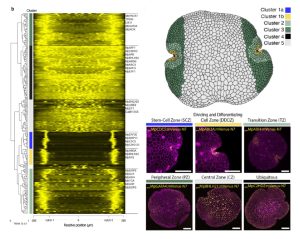 Marchantia’s power as a model organism continues to grow! Here, Ramoni et al. have investigated the expression pattern of the proximal promoters of most of its 450 transcription factor (TF)-encoding genes. The promoter elements were fused to nuclear-localized fluorescent reporters and introduced into plants. The authors provide the expression data of the constructs in a useful and accessible website, https://mpexpatdb.org/. Each TF promoter construct is shown along with chlorophyll fluorescence and a constitutive plasma membrane marker, which can be toggled on and off independently. Focusing on 218 genes, they identified five expression-pattern based clusters, with many of the genes preferentially expressed in the stem-cell zone or the dividing and differentiating cell zone. They also examined cell-specific and dynamic patterns of expression, and correlated these results with those obtained through other methods such as transcriptomics. As well as providing a great resource for further studies, this work has already provided insights into Marchantia development, including a suite of genes expressed in the stem-cell zone that are distinct from genes expressed in vascular plant meristems. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae053
Marchantia’s power as a model organism continues to grow! Here, Ramoni et al. have investigated the expression pattern of the proximal promoters of most of its 450 transcription factor (TF)-encoding genes. The promoter elements were fused to nuclear-localized fluorescent reporters and introduced into plants. The authors provide the expression data of the constructs in a useful and accessible website, https://mpexpatdb.org/. Each TF promoter construct is shown along with chlorophyll fluorescence and a constitutive plasma membrane marker, which can be toggled on and off independently. Focusing on 218 genes, they identified five expression-pattern based clusters, with many of the genes preferentially expressed in the stem-cell zone or the dividing and differentiating cell zone. They also examined cell-specific and dynamic patterns of expression, and correlated these results with those obtained through other methods such as transcriptomics. As well as providing a great resource for further studies, this work has already provided insights into Marchantia development, including a suite of genes expressed in the stem-cell zone that are distinct from genes expressed in vascular plant meristems. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1093/plcell/koae053
Novel lignin-based extracellular barrier in glandular trichome
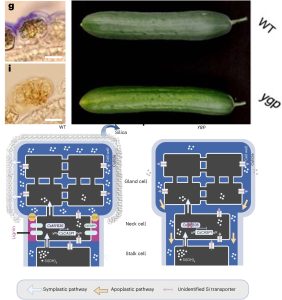 This is a fascinating story by Hao et al., which takes us from phenotype to novel insights by looking at glandular trichomes in cucumber. Cucumber fruits often have a silica-based white powder deposited on their surface, but this is missing from the yellow green peel (ygp) mutant. Staining shows that the silica is deposited on the surface of glandular trichomes of wild-type plants but not on the mutants. The YGP gene encodes MYB36, which in Arabidopsis is a master regulator of Casparian strip formation. The Casparian strip is a lignin-based apoplastic barrier that controls apoplastic movement in the root endodermis. Transcriptomic analysis showed that in the ygp mutant one of the most down-regulated genes is CASP1, which is a direct target of MYB26 and in the endodermis is a scaffold protein for Casparian strip formation. The authors looked at the expression pattern of CASP1 in cucumber and found that it is highly expressed in the neck cells of glandular trichomes. Additional experiments showed that in the neck region, there is an apoplastic accumulation of lignin as has been observed in the Casparian strip. The authors propose a model in which there is an apoplastic barrier in the neck region of glandular trichomes that keeps the silica exudate on the surface of these cells, but in the ypg mutant is missing, allowing this exudate to move apoplasticaly away from the surface. Finally, they identified similar structures of glandular trichomes of several other species. Given the usefulness of several compounds produced by glandular trichomes, it will be interesting to see if these new insights can be applied commercially. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01626-x
This is a fascinating story by Hao et al., which takes us from phenotype to novel insights by looking at glandular trichomes in cucumber. Cucumber fruits often have a silica-based white powder deposited on their surface, but this is missing from the yellow green peel (ygp) mutant. Staining shows that the silica is deposited on the surface of glandular trichomes of wild-type plants but not on the mutants. The YGP gene encodes MYB36, which in Arabidopsis is a master regulator of Casparian strip formation. The Casparian strip is a lignin-based apoplastic barrier that controls apoplastic movement in the root endodermis. Transcriptomic analysis showed that in the ygp mutant one of the most down-regulated genes is CASP1, which is a direct target of MYB26 and in the endodermis is a scaffold protein for Casparian strip formation. The authors looked at the expression pattern of CASP1 in cucumber and found that it is highly expressed in the neck cells of glandular trichomes. Additional experiments showed that in the neck region, there is an apoplastic accumulation of lignin as has been observed in the Casparian strip. The authors propose a model in which there is an apoplastic barrier in the neck region of glandular trichomes that keeps the silica exudate on the surface of these cells, but in the ypg mutant is missing, allowing this exudate to move apoplasticaly away from the surface. Finally, they identified similar structures of glandular trichomes of several other species. Given the usefulness of several compounds produced by glandular trichomes, it will be interesting to see if these new insights can be applied commercially. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01626-x
Using emmer wheat to discover genes involved in drought tolerance
 Wheat is a major global crop, but its yields are severely compromised by drought, thus developing varieties with a higher drought tolerance is important. Emmer wheat (Triticum turgidum) is a wheat ancestor that contains more genetic diversity than cultivated varieties, so it can be used in genome wide association studies (GWAS) to discover alleles associated with higher drought tolerance. Here Yang et al. grew 107 emmer wheat accessions to the three-leaf stage, and then transferred them to well-watered and water-stressed conditions for a further four weeks. For each accession, they measured biomass, root length, root area and root volume in both conditions and calculated drought tolerance indexes for each trait by dividing the value in the water-stressed condition by that in the well-watered condition. These drought tolerance indexes were used as input for a GWAS and an expression GWAS (eGWAS), which allows identification of loci associated with changes in gene expression between well-watered and water-stressed conditions. Integrating these strategies identified 86 quantitative trait loci (QTLs) and 190 expression QTLs. Interestingly, one contained the SUMO protease OTS1, and silencing or knocking this out in emmer wheat increased the drought tolerance indexes. Hence, OST1 has a potential to improve wheat drought tolerance. (Summary by Rose McNelly @Rose_McN) New Phytol. 10.1111/nph.19589
Wheat is a major global crop, but its yields are severely compromised by drought, thus developing varieties with a higher drought tolerance is important. Emmer wheat (Triticum turgidum) is a wheat ancestor that contains more genetic diversity than cultivated varieties, so it can be used in genome wide association studies (GWAS) to discover alleles associated with higher drought tolerance. Here Yang et al. grew 107 emmer wheat accessions to the three-leaf stage, and then transferred them to well-watered and water-stressed conditions for a further four weeks. For each accession, they measured biomass, root length, root area and root volume in both conditions and calculated drought tolerance indexes for each trait by dividing the value in the water-stressed condition by that in the well-watered condition. These drought tolerance indexes were used as input for a GWAS and an expression GWAS (eGWAS), which allows identification of loci associated with changes in gene expression between well-watered and water-stressed conditions. Integrating these strategies identified 86 quantitative trait loci (QTLs) and 190 expression QTLs. Interestingly, one contained the SUMO protease OTS1, and silencing or knocking this out in emmer wheat increased the drought tolerance indexes. Hence, OST1 has a potential to improve wheat drought tolerance. (Summary by Rose McNelly @Rose_McN) New Phytol. 10.1111/nph.19589
Peptide GOLVEN10 alters root development and noduletaxis
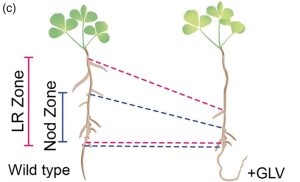 For many years, biologists argued about whether plants have peptide hormones like animals have, and ever since it was first shown that plant peptides do have hormone-like functions, I’ve said a little cheer as new functions are discovered (Yay plants!). A few years ago, Sonali Roy and colleagues wrote an excellent Teaching Tool on this topic (Small and mighty: Peptide hormones in plant biology) which I encourage you to read and share. Here, Roy et al. demonstrate a new role for the GOLVEN10 peptide in Medicago truncatula. Previous work identified roles for GOLVEN (GLV) peptides in root development; GLV overexpression leads to a wavy root and golven means “waves” in Dutch. The authors found that in Medicago, GLV10 is expressed in nodule-initiation sites and under the control of the NODULE INCEPTION (NIN) transcription factor. They found that application of GLV10 peptide to roots caused a decrease in nodule number, independent of effects on the symbiont. Interestingly, they also found a shift in position of the nodules and lateral roots along the root axis away from the root base, and that GLV10-treated roots had more, shorter cortical cells. Consistent with this finding, analysis of genes regulated by GLV10 showed that cell-cycle and cytoskeleton-related genes are differentially expressed. Thus, GLV10 is a regulator of nodule placement, or, by analogy to phyllotaxis, “nodultaxis”. (Summary by Mary Williams @PlantTeaching) Plant J. 10.1111/tpj.16626
For many years, biologists argued about whether plants have peptide hormones like animals have, and ever since it was first shown that plant peptides do have hormone-like functions, I’ve said a little cheer as new functions are discovered (Yay plants!). A few years ago, Sonali Roy and colleagues wrote an excellent Teaching Tool on this topic (Small and mighty: Peptide hormones in plant biology) which I encourage you to read and share. Here, Roy et al. demonstrate a new role for the GOLVEN10 peptide in Medicago truncatula. Previous work identified roles for GOLVEN (GLV) peptides in root development; GLV overexpression leads to a wavy root and golven means “waves” in Dutch. The authors found that in Medicago, GLV10 is expressed in nodule-initiation sites and under the control of the NODULE INCEPTION (NIN) transcription factor. They found that application of GLV10 peptide to roots caused a decrease in nodule number, independent of effects on the symbiont. Interestingly, they also found a shift in position of the nodules and lateral roots along the root axis away from the root base, and that GLV10-treated roots had more, shorter cortical cells. Consistent with this finding, analysis of genes regulated by GLV10 showed that cell-cycle and cytoskeleton-related genes are differentially expressed. Thus, GLV10 is a regulator of nodule placement, or, by analogy to phyllotaxis, “nodultaxis”. (Summary by Mary Williams @PlantTeaching) Plant J. 10.1111/tpj.16626
Cross-enrichment of-siderophore secreting rhizobacteria improves iron nutrition and yield of peanut intercropped with maize
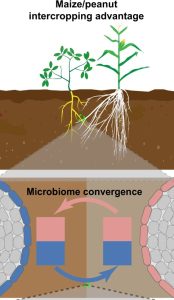 Intercropping, the system of growing at least two crops simultaneously, increases crop productivity and ecological sustainability. An intercropping system of peanut (Arachis hypogaea L.) and maize (Zea mays) has been previously found to improve the yield of peanut, specifically its iron nutrition and photosynthetic efficiency. Here, Wang et al. investigated the role of the rhizosphere microbiome in the increased iron nutrition observed in peanut/maize intercropping systems. They used 16S rRNA amplicon sequencing to explore the microbiome communities of peanut and maize in monocropping and intercropping systems. They found a convergence between maize and peanut microbiomes in the intercropping system. Interestingly, siderophore-producing Pseudomonas spp. was cross-enriched from maize to peanut and found to be associated with improved iron nutrition in peanut. The authors conclude that the improved yield of peanut observed in the peanut-maize intercropping system is associated with the cross-enrichment of rhizosphere microbiomes from maize. These findings highlight the potential role of rhizosphere microbiomes in intercropping systems and provide insights into the possibility of exploring such systems to boost crop yield and productivity. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Nature Comms. 10.1038/s41467-024-45207-0
Intercropping, the system of growing at least two crops simultaneously, increases crop productivity and ecological sustainability. An intercropping system of peanut (Arachis hypogaea L.) and maize (Zea mays) has been previously found to improve the yield of peanut, specifically its iron nutrition and photosynthetic efficiency. Here, Wang et al. investigated the role of the rhizosphere microbiome in the increased iron nutrition observed in peanut/maize intercropping systems. They used 16S rRNA amplicon sequencing to explore the microbiome communities of peanut and maize in monocropping and intercropping systems. They found a convergence between maize and peanut microbiomes in the intercropping system. Interestingly, siderophore-producing Pseudomonas spp. was cross-enriched from maize to peanut and found to be associated with improved iron nutrition in peanut. The authors conclude that the improved yield of peanut observed in the peanut-maize intercropping system is associated with the cross-enrichment of rhizosphere microbiomes from maize. These findings highlight the potential role of rhizosphere microbiomes in intercropping systems and provide insights into the possibility of exploring such systems to boost crop yield and productivity. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Nature Comms. 10.1038/s41467-024-45207-0
Application of silicate enhances plant growth-promoting rhizobacteria network complexity in sugarcane rhizosphere
Silicon (Si) plays a significant role in helping plants to mitigate biotic and abiotic stress. Previous studies have shown that the application of S i also influences the microbial communities in the rhizosphere. However, the extent to which Si shapes the dynamics of plant growth promoting rhizobacteria (PGPR) in the rhizosphere is less explored. Here, Leite et al. investigated the influence of silicon on the PGPR community in the sugarcane rhizosphere. They assessed the PGPR community within the rhizosphere of three sugarcane genotypes in the presence (Si+) and absence (Si-) of Si in the form of silicate. Specifically, the authors used 16S rRNA sequencing to evaluate the richness, diversity, composition, co-occurrence network, and niche occupancy of the PGPR community. Although the results showed no significant influence of the treatments and the genotypes on the richness and diversity of the PGPR, they found varied effects of the Si treatment across the three genotypes. Two of the three genotypes showed increased network complexity in the presence of Si, indicating that the effect of the Si treatment may be genotype dependent. The authors concluded that Si application increases the complexity of microbial networks with the potential to increase plant growth. These findings highlight the possibility of increasing PGPR interaction networks as a recipe for sustainable agriculture. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Rhizosphere 10.1016/j.rhisph.2024.100855
i also influences the microbial communities in the rhizosphere. However, the extent to which Si shapes the dynamics of plant growth promoting rhizobacteria (PGPR) in the rhizosphere is less explored. Here, Leite et al. investigated the influence of silicon on the PGPR community in the sugarcane rhizosphere. They assessed the PGPR community within the rhizosphere of three sugarcane genotypes in the presence (Si+) and absence (Si-) of Si in the form of silicate. Specifically, the authors used 16S rRNA sequencing to evaluate the richness, diversity, composition, co-occurrence network, and niche occupancy of the PGPR community. Although the results showed no significant influence of the treatments and the genotypes on the richness and diversity of the PGPR, they found varied effects of the Si treatment across the three genotypes. Two of the three genotypes showed increased network complexity in the presence of Si, indicating that the effect of the Si treatment may be genotype dependent. The authors concluded that Si application increases the complexity of microbial networks with the potential to increase plant growth. These findings highlight the possibility of increasing PGPR interaction networks as a recipe for sustainable agriculture. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Rhizosphere 10.1016/j.rhisph.2024.100855
Effect of plant-derived versus animal-derived fertilizers on the rhizosphere microbiome
 Yu et al. conducted a meta-analysis of published 16S rRNA gene amplicon sequenced soil samples to compare the effects of plant-based fertilizers (e.g. compost, seaweed fertilizer) versus animal-based fertilizers (e.g., dung, manure) versus control, unfertilized samples on soil microbial diversity and function. Their analysis showed that the fertilizer source significantly influences microbial functional profiles and the metabolic pathways and biogeochemical cycle processes that are upregulated. The authors validated these findings experimentally by investigating the effect of the four fertilizer sources (fermented pig or chicken manure, fermented tree leaves or tea slag) on pakchoi (Brassica chinensis L.) All fertilizers showed a significant increase in plant growth versus unfertilized treatment and confirmed the findings of their meta-analysis on the impact of soil microbial function. Both types of fertilizers increased the stability of rhizosphere microbial communities, but in different ways. The animal-derived fertilizer mainly enriched genes related to nitrogen cycling while the plant-derived fertilizer enriched genes related to carbon fixation, carbon degradation, and methane metabolism. However, in samples treated with animal-derived fertilizers there was an increased abundance of antibiotic resistance genes and viruses related to human disease. Given the risks to human health conferred by animal-derived fertilizer samples, the authors conclude that plant-derived fertilizers are safer for sustainable crop production. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Appl. Environ. Microbiol. 10.1128/aem.01719-23
Yu et al. conducted a meta-analysis of published 16S rRNA gene amplicon sequenced soil samples to compare the effects of plant-based fertilizers (e.g. compost, seaweed fertilizer) versus animal-based fertilizers (e.g., dung, manure) versus control, unfertilized samples on soil microbial diversity and function. Their analysis showed that the fertilizer source significantly influences microbial functional profiles and the metabolic pathways and biogeochemical cycle processes that are upregulated. The authors validated these findings experimentally by investigating the effect of the four fertilizer sources (fermented pig or chicken manure, fermented tree leaves or tea slag) on pakchoi (Brassica chinensis L.) All fertilizers showed a significant increase in plant growth versus unfertilized treatment and confirmed the findings of their meta-analysis on the impact of soil microbial function. Both types of fertilizers increased the stability of rhizosphere microbial communities, but in different ways. The animal-derived fertilizer mainly enriched genes related to nitrogen cycling while the plant-derived fertilizer enriched genes related to carbon fixation, carbon degradation, and methane metabolism. However, in samples treated with animal-derived fertilizers there was an increased abundance of antibiotic resistance genes and viruses related to human disease. Given the risks to human health conferred by animal-derived fertilizer samples, the authors conclude that plant-derived fertilizers are safer for sustainable crop production. (Summary by Abdulkabir Omeiza Abdulmalik @Omeiza_PlantDoc) Appl. Environ. Microbiol. 10.1128/aem.01719-23



