Plant Science Research Weekly: June 7th
Review: Source–sink regulation in crops under water deficit ($)
 Plants have a remarkable ability to coordinate cellular activities across huge distances, yet we have only a basic understanding of how these remote activities are coordinated. A review by Rodrigues et al. summarizes what we know about the relationship between source (e.g., photosynthetic tissues) and sinks (e.g., growing tissues or seeds), and particularly how this relationship is affected by water deficit (WD). As the authors describe, greater insights into whole-plant physiological integration is needed in order to develop high-yielding, drought-tolerant crops. The authors summarize how both source and sink activities are affected by WD, at physiological as well as cellular and biochemical levels. As examples, WD closes stomata and reduces gas exchange, leading to a decrease in photosynthetic carbon fixation. WD also decrease xylem flow, causing also a decrease in phloem flow and the movement of sugars into the sink tissues. The authors discuss results from genetic studies that reveal the contributions of specific transcription factors and hormones, as well as the metabolic integrators SnRK1 and TOR. Finally, they describe the many questions that need additional research. This excellent, accessible review would be useful for students to help them integrate molecular and physiological stress responses. (Summary by Mary Williams) Trends Plant Sci 10.1016/j.tplants.2019.04.005
Plants have a remarkable ability to coordinate cellular activities across huge distances, yet we have only a basic understanding of how these remote activities are coordinated. A review by Rodrigues et al. summarizes what we know about the relationship between source (e.g., photosynthetic tissues) and sinks (e.g., growing tissues or seeds), and particularly how this relationship is affected by water deficit (WD). As the authors describe, greater insights into whole-plant physiological integration is needed in order to develop high-yielding, drought-tolerant crops. The authors summarize how both source and sink activities are affected by WD, at physiological as well as cellular and biochemical levels. As examples, WD closes stomata and reduces gas exchange, leading to a decrease in photosynthetic carbon fixation. WD also decrease xylem flow, causing also a decrease in phloem flow and the movement of sugars into the sink tissues. The authors discuss results from genetic studies that reveal the contributions of specific transcription factors and hormones, as well as the metabolic integrators SnRK1 and TOR. Finally, they describe the many questions that need additional research. This excellent, accessible review would be useful for students to help them integrate molecular and physiological stress responses. (Summary by Mary Williams) Trends Plant Sci 10.1016/j.tplants.2019.04.005
Insight: Rapid evolution in plant – microbe interactions
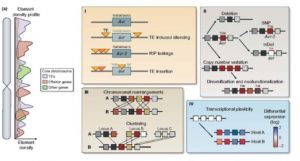 Plants and associated microbes co-exist and co-evolve over time. The rate of evolution is higher in pathogens as compared to plants. Although the phenotypic consequences of rapid evolution in pathogens are well studied, changes at genomic level are not as well known, and are the subject of a recent review by Frantzaskakis et al. Duplication and diversion of plant R genes, accompanied by evolution of pathogen Avr genes, are “testimonies of an ongoing arms race”. Genomic modifications by processes such as deletions, TE insertions, and epigenetic gene silencing of avirulence genes can lead to increased pathogen virulence. All these genomic changes together contribute to the ability of a microbe to become established as a pathogen in spite of the plant’s molecular warfare. Additional complexity is conferred by the recently discovered transfer of small RNAs from pathogen to plant, which can increase virulence. This review also describes the contributions of of transcriptional plasticity and gene dosage to the plant-pathogen battlefield. (Summary by Mugdha Sabale) New Phytol. 10.1111/nph.15966
Plants and associated microbes co-exist and co-evolve over time. The rate of evolution is higher in pathogens as compared to plants. Although the phenotypic consequences of rapid evolution in pathogens are well studied, changes at genomic level are not as well known, and are the subject of a recent review by Frantzaskakis et al. Duplication and diversion of plant R genes, accompanied by evolution of pathogen Avr genes, are “testimonies of an ongoing arms race”. Genomic modifications by processes such as deletions, TE insertions, and epigenetic gene silencing of avirulence genes can lead to increased pathogen virulence. All these genomic changes together contribute to the ability of a microbe to become established as a pathogen in spite of the plant’s molecular warfare. Additional complexity is conferred by the recently discovered transfer of small RNAs from pathogen to plant, which can increase virulence. This review also describes the contributions of of transcriptional plasticity and gene dosage to the plant-pathogen battlefield. (Summary by Mugdha Sabale) New Phytol. 10.1111/nph.15966
Designer pentatricopeptide repeat PPR protein as RNA affinity tag
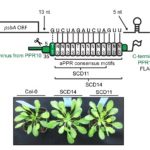
 Pentatricopeptide repeat (PPR) proteins bind RNA, using a code in which specific amino acid motifs recognize specific bases, similar to TAL effector proteins. In plants, PPR proteins localize almost exclusively to mitochondria and chloroplasts, with functions including RNA, stabilization, splicing and editing. Here, McDermott et al. used insights about the amino-acid to RNA code to design artificial PPR (aPPRs) proteins that bind target RNAs with high specificity in vivo. These aPPRs can then be used to isolate specific ribonucleoprotein particles (RNPs; associations of RNA + proteins) to characterize their constituents. The authors designed an aPPR that specifically binds the 3’ UTR of the chloroplast psbA mRNA, which encodes photosystem II protein D1. Their study identified psbA mRNA binding proteins of unknown function, and also demonstrated the feasibility of using aPPRs to characterize proteins that bind to specific RNAs. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00177
Pentatricopeptide repeat (PPR) proteins bind RNA, using a code in which specific amino acid motifs recognize specific bases, similar to TAL effector proteins. In plants, PPR proteins localize almost exclusively to mitochondria and chloroplasts, with functions including RNA, stabilization, splicing and editing. Here, McDermott et al. used insights about the amino-acid to RNA code to design artificial PPR (aPPRs) proteins that bind target RNAs with high specificity in vivo. These aPPRs can then be used to isolate specific ribonucleoprotein particles (RNPs; associations of RNA + proteins) to characterize their constituents. The authors designed an aPPR that specifically binds the 3’ UTR of the chloroplast psbA mRNA, which encodes photosystem II protein D1. Their study identified psbA mRNA binding proteins of unknown function, and also demonstrated the feasibility of using aPPRs to characterize proteins that bind to specific RNAs. (Summary by Mary Williams) Plant Cell 10.1105/tpc.19.00177
Gene networks underlying cannabinoid and terpenoid accumulation in cannabis
 Recent changes in the legal status of cannabis and cannabis-products have allowed renewed scientific interest in the genetic and metabolic basis of cannabis’ effects as well as the regulatory issues related to quality control of the products. Many varieties of Cannabis spp. have poorly documented pedigrees, and a focus on breeding simply for increased THC has left the chemical diversity relatively understudied. In this recent study, Zager et al. have characterized nine varieties of hybrid Cannabis based on their 1) metabolite profiles for a variety of psychoactive or fragrant compounds, and 2) gene expression profiles in the glandular hairs known to by the site of synthesis for many of these compounds. The authors use several dimensional reduction and clustering techniques to confirm that the Cannabis varieties have low within-variety variability and also cluster according to purported species dominance in the hybrid cross (C. sativa or C. indica). Having established a strong gene-to-metabolite correlation, the authors go on to functionally characterize several enzymes in the cannabinoid/terpenoid biosynthesis pathway. These results lay a foundation for further metabolic studies of this valuable plant, and also provide a framework for verification Cannabis metabolite content in a commercial setting. (Summary by Alex Rajewski) Plant Phys 10.1104/pp.18.01506
Recent changes in the legal status of cannabis and cannabis-products have allowed renewed scientific interest in the genetic and metabolic basis of cannabis’ effects as well as the regulatory issues related to quality control of the products. Many varieties of Cannabis spp. have poorly documented pedigrees, and a focus on breeding simply for increased THC has left the chemical diversity relatively understudied. In this recent study, Zager et al. have characterized nine varieties of hybrid Cannabis based on their 1) metabolite profiles for a variety of psychoactive or fragrant compounds, and 2) gene expression profiles in the glandular hairs known to by the site of synthesis for many of these compounds. The authors use several dimensional reduction and clustering techniques to confirm that the Cannabis varieties have low within-variety variability and also cluster according to purported species dominance in the hybrid cross (C. sativa or C. indica). Having established a strong gene-to-metabolite correlation, the authors go on to functionally characterize several enzymes in the cannabinoid/terpenoid biosynthesis pathway. These results lay a foundation for further metabolic studies of this valuable plant, and also provide a framework for verification Cannabis metabolite content in a commercial setting. (Summary by Alex Rajewski) Plant Phys 10.1104/pp.18.01506
Embryo protection after germination in date palm
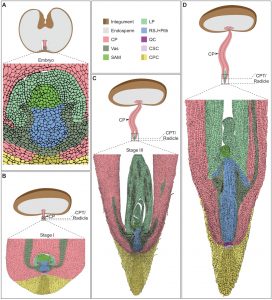 Plant morphogenesis is a dynamic process that can be modulated in response to environmental cues. In this work, Xiao et al. characterized germination and seedling development in date palm. After germination, the cotyledonary petiole grows, but the embryo development is paused. At early stages of development a low level of cell division in the embryo is observed, but cell division frequency increases at later stages. The early growth arrest in the embryo is correlated with high levels of abscisic acid (ABA); on the other hand, in the active tissues such as the cotyledonary petiole, low levels of ABA and high levels of gibberellic acid (GA) were detected. About three weeks after germination, embryo growth is resumed and its development continues inside the cotyledonary petiole that acts as a shield. The encapsulated seedling displays root and shoot features, for instance, accumulation of starch granules and transcripts of auxin response genes (i.e. PdIAA2) in the root tip, and, the expression of the SHOOT MERISTEMLESS ortholog gen (PdSTM) in the SAM. The authors also characterized the date palm root system architecture non-invasively using X-ray micro-computed tomography. Histochemical analysis of roots revealed accumulation of suberin and lignin, which are related to water loss prevention. Understanding these and other adaptations to extreme environments in the desert plants could aid crop improvement. (Summary by Humberto Herrera-Ubaldo) Plant Cell 10.1105/tpc.19.00008
Plant morphogenesis is a dynamic process that can be modulated in response to environmental cues. In this work, Xiao et al. characterized germination and seedling development in date palm. After germination, the cotyledonary petiole grows, but the embryo development is paused. At early stages of development a low level of cell division in the embryo is observed, but cell division frequency increases at later stages. The early growth arrest in the embryo is correlated with high levels of abscisic acid (ABA); on the other hand, in the active tissues such as the cotyledonary petiole, low levels of ABA and high levels of gibberellic acid (GA) were detected. About three weeks after germination, embryo growth is resumed and its development continues inside the cotyledonary petiole that acts as a shield. The encapsulated seedling displays root and shoot features, for instance, accumulation of starch granules and transcripts of auxin response genes (i.e. PdIAA2) in the root tip, and, the expression of the SHOOT MERISTEMLESS ortholog gen (PdSTM) in the SAM. The authors also characterized the date palm root system architecture non-invasively using X-ray micro-computed tomography. Histochemical analysis of roots revealed accumulation of suberin and lignin, which are related to water loss prevention. Understanding these and other adaptations to extreme environments in the desert plants could aid crop improvement. (Summary by Humberto Herrera-Ubaldo) Plant Cell 10.1105/tpc.19.00008
Alternative usage of miRNA-biogenesis co-factors in plants at low temperatures
 As in animals, plants produce microRNAs that are also key developmental regulators. Unlike some animals, plants are more exposed to environmental factors that alter cellular processes. Ré et al. show here that phenotypic defects produced by loss-of-function mutations in key proteins of Arabidopsis miRNA biogenesis machinery, such as SERRATE (SE) and HYPONASTIC LEAVES 1 (HYL1), are milder at low temperature (16°C vs 22°C). They show that at this temperature, the production of several miRNA is partially restored. Moreover, in a loss-of-function allele of the HYL1 cofactor, they identified that specific miRNA are either dependent or independent of this cofactor at low temperatures, where the ribonuclease excision step precision in pri-miRNA maturation is improved at low temperatures in the firsts. Interestingly miRNAs affected by temperature and HYL1 poses specific features related to their secondary structure. This suggests that low temperature could be helping to stabilize miRNA secondary structure, making cofactors more dispensable. Authors also discuss how this regulation could be evolutionarily relevant for environmental adaptation, for example, during the day-night temperature shift. This paper highlights that miRNA biogenesis is not constant and ubiquitous as previously thought, and open questions on how the environment influences these processes. (Summary by Facundo Romani) Development 10.1242/dev.172932
As in animals, plants produce microRNAs that are also key developmental regulators. Unlike some animals, plants are more exposed to environmental factors that alter cellular processes. Ré et al. show here that phenotypic defects produced by loss-of-function mutations in key proteins of Arabidopsis miRNA biogenesis machinery, such as SERRATE (SE) and HYPONASTIC LEAVES 1 (HYL1), are milder at low temperature (16°C vs 22°C). They show that at this temperature, the production of several miRNA is partially restored. Moreover, in a loss-of-function allele of the HYL1 cofactor, they identified that specific miRNA are either dependent or independent of this cofactor at low temperatures, where the ribonuclease excision step precision in pri-miRNA maturation is improved at low temperatures in the firsts. Interestingly miRNAs affected by temperature and HYL1 poses specific features related to their secondary structure. This suggests that low temperature could be helping to stabilize miRNA secondary structure, making cofactors more dispensable. Authors also discuss how this regulation could be evolutionarily relevant for environmental adaptation, for example, during the day-night temperature shift. This paper highlights that miRNA biogenesis is not constant and ubiquitous as previously thought, and open questions on how the environment influences these processes. (Summary by Facundo Romani) Development 10.1242/dev.172932
Auxin-mediated statolith production for root gravitropism
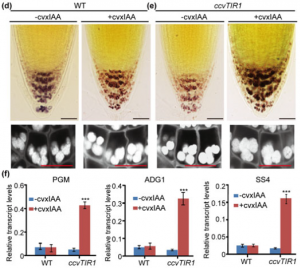 Plants have evolved the ability to perceive and respond to gravity, one of the most persistent external cues. This response process, termed gravitropism, has been extensively studied for its importance in plant development and its potential to improve agronomic traits of crops. The gravitropic response of plants is defined by three main steps: sedimentation of starch-filled amyloplasts within gravity sensing cells, the signal transduction of that sedimentation event, and asymmetric auxin distribution followed by differential growth. Zhang et al. probed auxin’s influence on the size and number of amyloplasts within the root cap of Arabidopsis thaliana. The group found that application of auxin analogues increased amyloplast accumulation and upregulated starch synthesis genes, while auxin synthesis inhibitors achieved the opposite result. Knockout mutations of auxin biosynthesis genes showed decreased amyloplast accumulation and starch synthesis gene upregulation, while mutations in auxin transport genes increased these same outputs. Furthermore, starch synthesis genes were shown to be auxin-regulated through mutations in auxin signaling pathway genes and the use of an inducible TIR1 complex. These results show that auxin may play an important role in helping plants sense as well as respond to gravity. (Summary by Nathan Scinto-Madonich) New Phytol. 10.1111/nph.15932
Plants have evolved the ability to perceive and respond to gravity, one of the most persistent external cues. This response process, termed gravitropism, has been extensively studied for its importance in plant development and its potential to improve agronomic traits of crops. The gravitropic response of plants is defined by three main steps: sedimentation of starch-filled amyloplasts within gravity sensing cells, the signal transduction of that sedimentation event, and asymmetric auxin distribution followed by differential growth. Zhang et al. probed auxin’s influence on the size and number of amyloplasts within the root cap of Arabidopsis thaliana. The group found that application of auxin analogues increased amyloplast accumulation and upregulated starch synthesis genes, while auxin synthesis inhibitors achieved the opposite result. Knockout mutations of auxin biosynthesis genes showed decreased amyloplast accumulation and starch synthesis gene upregulation, while mutations in auxin transport genes increased these same outputs. Furthermore, starch synthesis genes were shown to be auxin-regulated through mutations in auxin signaling pathway genes and the use of an inducible TIR1 complex. These results show that auxin may play an important role in helping plants sense as well as respond to gravity. (Summary by Nathan Scinto-Madonich) New Phytol. 10.1111/nph.15932
Genetic and Molecular analysis of trichome development in Arabis alpina $
 Trichomes (plant hairs), arise from epidermal cells of plants. The molecular mechanism involved in the development of trichomes of plants has been well studied in Arabidopsis thaliana. In this paper, Chopra et al. have identified key regulators of leaf trichome development in Arabis alpina, a species that diverged from A. thaliana about 26-40 million years ago. Using forward genetics, multiple regulators of trichome development were identified in A. alpina including mutants that cause a glabrous (trichome lacking) phenotype. Although most of the genes involved in trichome development of A. alpina were similar to those in A. thaliana, some regulators have diversified in their function. One of the major findings concerns the role of GL3 (GLABRA3), which when mutated causes a strong phenotype in A. alpina but requires simultaneous mutation in EGL3 (ENHANCER OF GLABRA3) to cause a strong glabrous phenotype in A. thaliana. Overall the results from this paper suggests that with close relationship between species, gene functions are comparable but the divergence is sufficient to study some difference in some molecular players of development. (Summary by Suresh Damodaran) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1819440116
Trichomes (plant hairs), arise from epidermal cells of plants. The molecular mechanism involved in the development of trichomes of plants has been well studied in Arabidopsis thaliana. In this paper, Chopra et al. have identified key regulators of leaf trichome development in Arabis alpina, a species that diverged from A. thaliana about 26-40 million years ago. Using forward genetics, multiple regulators of trichome development were identified in A. alpina including mutants that cause a glabrous (trichome lacking) phenotype. Although most of the genes involved in trichome development of A. alpina were similar to those in A. thaliana, some regulators have diversified in their function. One of the major findings concerns the role of GL3 (GLABRA3), which when mutated causes a strong phenotype in A. alpina but requires simultaneous mutation in EGL3 (ENHANCER OF GLABRA3) to cause a strong glabrous phenotype in A. thaliana. Overall the results from this paper suggests that with close relationship between species, gene functions are comparable but the divergence is sufficient to study some difference in some molecular players of development. (Summary by Suresh Damodaran) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1819440116
A fine-scale genetic linkage map reveals genomic regions associated with economic traits in walnut (Juglans regia)
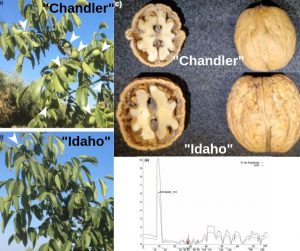 The bearing habit refers to the location of flower buds in woody plants. For walnut cultivars, lateral bearing is preferred for its yield superiority. Deciphering its genetic architecture would improve seedling and sapling selection. Here, Aradhya et al. performed a SNP-based linkage map to estimate genetic effects of quantitative trait loci (QTLs) for 5 yield attributes in the Persian walnut (Juglans regia L.): lateral bearing, harvest date, shell thickness, nut weight and kernel fill. Using phenotype data from F1 trees from contrasting heterozygous genotypes, “Chandler” (lateral bearing) x “Idaho” (terminal bearing). “Chandler”-based SNP array was used to mapped 2,220 SNPs markers into 16 linkage groups (LG). By a two-step strategy major effect QTLs were located, single QTL model followed by multiple QTL model (MQM), which uses neighbouring linked markers as cofactors reducing residual variation. For lateral bearing expression, a single major QTL in LG 11 showed negative additive effects, inferring its heterozygote superiority. Harvest time and all three nut traits appeared to be under the control of linked pleiotropic QTLs on LG 1 with marginal additive effects. (Summary by Ana Valladares). Plant Breeding. 10.1111/pbr.12703
The bearing habit refers to the location of flower buds in woody plants. For walnut cultivars, lateral bearing is preferred for its yield superiority. Deciphering its genetic architecture would improve seedling and sapling selection. Here, Aradhya et al. performed a SNP-based linkage map to estimate genetic effects of quantitative trait loci (QTLs) for 5 yield attributes in the Persian walnut (Juglans regia L.): lateral bearing, harvest date, shell thickness, nut weight and kernel fill. Using phenotype data from F1 trees from contrasting heterozygous genotypes, “Chandler” (lateral bearing) x “Idaho” (terminal bearing). “Chandler”-based SNP array was used to mapped 2,220 SNPs markers into 16 linkage groups (LG). By a two-step strategy major effect QTLs were located, single QTL model followed by multiple QTL model (MQM), which uses neighbouring linked markers as cofactors reducing residual variation. For lateral bearing expression, a single major QTL in LG 11 showed negative additive effects, inferring its heterozygote superiority. Harvest time and all three nut traits appeared to be under the control of linked pleiotropic QTLs on LG 1 with marginal additive effects. (Summary by Ana Valladares). Plant Breeding. 10.1111/pbr.12703
Nitrate–NRT1.1B–SPX4 cascade integrates N and P signaling networks
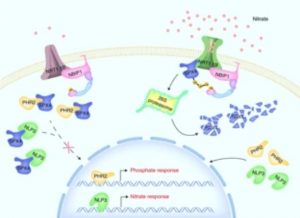 Plants need several mineral nutrients for their optimal growth and development. This is contingent on a proper nutrient balance particularly between the two most required essential elements, nitrogen (N) and phosphorus. Even though considerable success has been achieved in understanding the molecular mechanism mediating individual N and P signaling, the direct cross talk between N and P remain unknown. Hu and colleagues investigated the signaling network regulating the interaction between N and P in rice. Previously, NRT1.1B was shown to be a transceptor, sensing nitrate signal and activating downstream responses. Here, the authors show that nitrate activation of phosphate signaling is dependent on NRT1.1B. SPX4 is a repressor preventing the nuclear localization of PHR2 the master regulator of phosphate signaling in rice. Hu et al. showed that protein accumulation of SPX4 was decreased by nitrate induction of phosphate signaling via degradation by ubiquitination and there was a shift in PHR2 localization from the cytosol to the nucleus. This observation was further supported by gene expression analysis where nitrate-induced phosphate signaling was repressed by overexpression of SPX4 and phr2 mutant. The authors also showed that NPL3 the central regulator of nitrate signaling is also controlled by SPX4, indicating that NRT1.1B-SPX4 signaling cascade coordinates both nitrate and phosphate signaling. (Summary by Toluwase Olukayode) Nature Plants 10.1038/s41477-019-0384-1
Plants need several mineral nutrients for their optimal growth and development. This is contingent on a proper nutrient balance particularly between the two most required essential elements, nitrogen (N) and phosphorus. Even though considerable success has been achieved in understanding the molecular mechanism mediating individual N and P signaling, the direct cross talk between N and P remain unknown. Hu and colleagues investigated the signaling network regulating the interaction between N and P in rice. Previously, NRT1.1B was shown to be a transceptor, sensing nitrate signal and activating downstream responses. Here, the authors show that nitrate activation of phosphate signaling is dependent on NRT1.1B. SPX4 is a repressor preventing the nuclear localization of PHR2 the master regulator of phosphate signaling in rice. Hu et al. showed that protein accumulation of SPX4 was decreased by nitrate induction of phosphate signaling via degradation by ubiquitination and there was a shift in PHR2 localization from the cytosol to the nucleus. This observation was further supported by gene expression analysis where nitrate-induced phosphate signaling was repressed by overexpression of SPX4 and phr2 mutant. The authors also showed that NPL3 the central regulator of nitrate signaling is also controlled by SPX4, indicating that NRT1.1B-SPX4 signaling cascade coordinates both nitrate and phosphate signaling. (Summary by Toluwase Olukayode) Nature Plants 10.1038/s41477-019-0384-1