Plant Science Research Weekly: June 14, 2024
Review: Deep learning approaches to understanding stomatal function
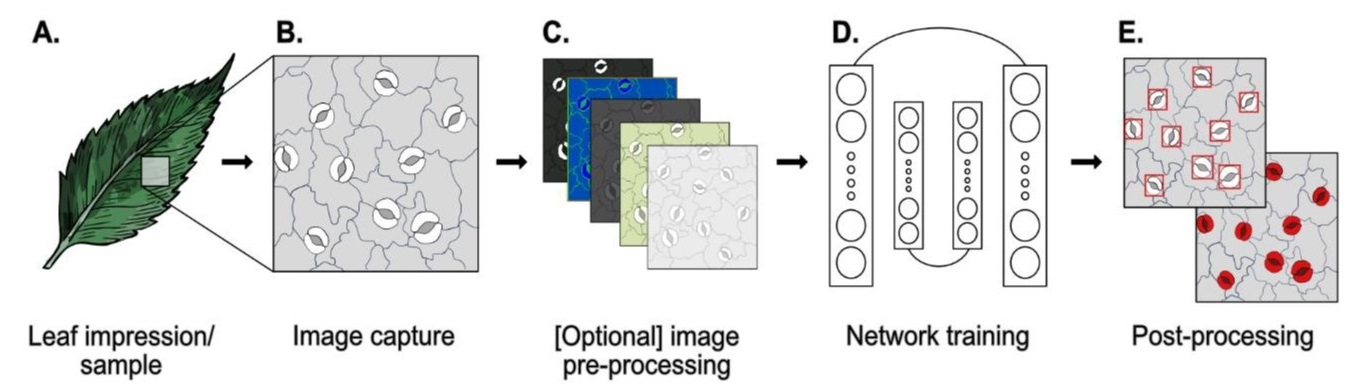
Sydney Brenner famously said, “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Right now, we’re seeing how advancements and new techniques in artificial intelligence and deep learning are being applied in plant sciences, including in the analysis of all sorts of large -omics datasets. This review by Gibbs and Burgess discusses deep learning approaches to understand stomatal function, and it provides a good overview of what is inside the (to me) black box of deep learning. Stomatal function depends on stomatal morphology, largely determined during development, and the very dynamic stomatal aperture, which responds to environment and physiology. The authors explain the steps in acquiring data, building models, and testing the models, and summarize methods and limitations for the study of stomata. They call for greater communication amongst those developing deep learning pipelines and greater transparency and sharing of datasets, and to encourage such collaboration and sharing they have created StomataHub (http://www.stomatahub.com/). Finally, they note that knowledge is currently poorly translated to field studies, which are integral for yield improvement, but that such translation is needed to generate new discoveries and ideas. (Summary by Mary Williams @PlantTeaching) J Exp Bot 10.1093/jxb/erae207
Review: Engineering plant–microbe communication for plant nutrient use efficiency
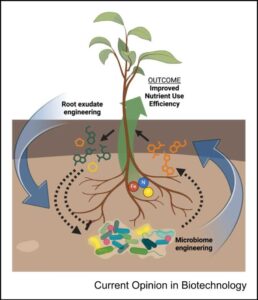
Plant nutrient use efficiency (NUE) has become a major concern in recent years as farmers and scientists strive to make agriculture more sustainable. To this end, the manipulation of plant-microbe interactions holds great potential. In this short review, Griffin et al. highlight recent findings, focusing on two approaches aimed at increasing NUE by acting on the rhizosphere environment: the engineering of plant exudates from roots to favor the growth of beneficial microbes, and the engineering of microbial communities (bioinoculants) to increase nutrient availability. Along with proof-of-concept studies to increase NUE, the authors discuss the importance of biocontainment strategies for the deployment of such technologies in the field. There is indeed a growing need to translate these kind of fundamental discoveries from a laboratory setting to the field, a process that is both technically and legally challenging. Future efforts based on recent advances in computational and synthetic biology techniques will hopefully pave the way for better NUE in a broad range of crops. (Summary by Carlo Pasini @Crl_Psn) Curr. Opin. Biotech. 10.1016/j.copbio.2024.103150
Flip the light switch for protein production
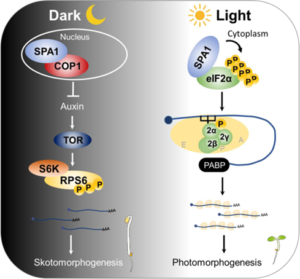
Protein synthesis, modification, and degradation are not only crucial for responding to environmental change, but they also represent significant energy costs for a plant. Energy homeostasis is highly affected by the presence of light. In young seedlings, light enhances translation during the dark-to-light transition, although we do not fully understand how this process is regulated. SPA kinases work with partners inside or outside the nuclei to trigger translation for skotomorphogenesis or photomorphogenesis. In mammalian and yeast cells, the reversible phosphorylation of eukaryotic initiation factor 2 alpha (eIF2α) at serine 51 regulates translation, but plants show exceptions. Recent research by Chang et al. reveals that light exposure prompts SPA kinases to phosphorylate the eIF2α C-terminus, excluding serine 51. This non-traditional phosphorylation enhances the translation of mRNAs involved in light responses, photosynthesis, and chlorophyll biosynthesis. (Summary by Yueh Cho @YuehCho1984) Nature Comms 10.1038/s41467-024-47848-7
Tailored protein stability for stress resilience in light and dark
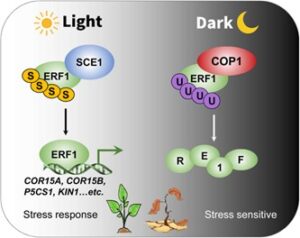
Light influences essentially all aspects of plant growth and development. Furthermore, plants are likely to adjust their stress responses based on fluctuations in energy availability during the dark-light cycle. Lin et al investigated a hormone downstream component, ETHYLENE RESPONSE FACTOR 1 (ERF1), and discovered that its posttranslational modifications are coordinated by dark or light conditions. In the light, sumoylated ERF1 is associated with SUMO‐CONJUGATING ENZYME 1 (SCE1) and triggers stress-responsive gene expression. On the other hand, CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) orchestrates ERF1 degradation in darkness by facilitating ubiquitination, negatively impacting stress tolerance mechanisms. This work sheds light on how plants integrate light signals and manage stress responses. (Summary by Yueh Cho @YuehCho1984) Plant Cell Environ. 10.1111/pce.14850.
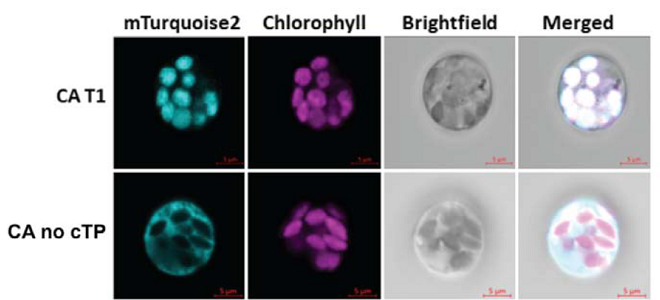
Engineering cytosolic carbonic anhydrase to establish C4 photosynthesis in rice
In mesophyll cells, carbonic anhydrase is mainly located in the chloroplast, however it is in the cytosol in plants with a C4 carbon concentrating mechanism. There is interest in relocating carbonic anhydrase to the cytosol of C3 plants as a first step in the introduction of a carbon concentrating mechanism. Here, Jethva et al. used CRISPR/Cas9 to generate rice (Oryza sativa) plants with cytosolic-localized carbonic anhydrase. They generated a mutant with a frame shift that disrupts the chloroplast transit peptide, however, to ensure the rest of the protein is produced correctly, the reading frame was restored at the end of the chloroplast transit peptide sequence. Carbonic anhydrase activity assays revealed that the mutants have 17-30% of the carbonic anhydrase activity compared to wild type plants, but crucially all of this is in the cytosol, with no chloroplastic activity detected. Despite the reduction in enzymatic activity, the plants have no abnormal growth or yield phenotype. Hence, it is possible to relocate carbonic anhydrase to the cytosol of mesophyll cells in rice, which could be a key step for the introduction of a complete carbon concentration mechanism. (Summary by Rose McNelly @Rose_McN) bioRxiv https://doi.org/10.1101/2024.05.21.595093
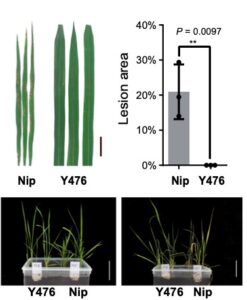
Crossing the gap: Decrypting the genome facilitates gene identification in wild rice
Domesticated crops provide a reliable food source but generally have little genetic diversity to cope with environmental fluctuations. By decrypting the genome of wild species such as the wild rice Oryza rufipogon, we may identify additional genetic variation useful for breeding process. However, the genomic heterozygosity of wild species is very challenging to work with. To cross this information gap, Huang et al. performed gapless assembly and comprehensive annotation of the wild rice Y476 genome. They incorporated segments of chromosomes from Y476 into the cultivated rice variety Nipponbare, allowing them to identify 254 Quantitative Trait Loci (QTLs) linked to agronomic traits and stress responses. Notably, a receptor-like kinase gene enhancing rice blast resistance was cloned from wild rice. This study provides a high-quality reference genome and establishes a platform for identifying beneficial genes in wild rice. (Summary by Yueh Cho @YuehCho1984) Nature Comms. 10.1038/s41467-024-48845-6.
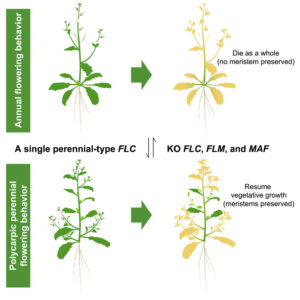
A single gene converts Brassicaceae from annual to perennial form
Polycarpic perennials, as opposed to annual plants, flower multiple times over several growing seasons throughout their lifecycle. To do that, they must retain a vegetative meristem that does not produce flowers during the reproductive phase. MADS-box transcription factors like FLOWERING LOCUS C (FLC) contribute to this process by repressing the floral transition in the meristem. While evidence suggests that annual plants in the Brassicaceae have evolved multiple times from perennial ancestors via changes in the expression pattern of FLC, the genetic details of these evolutionary transitions remain unclear. In this paper, Zhai and colleagues shed light on the molecular mechanisms that allow the switch from a perennial to an annual lifestyle within the Brassicaceae, by combining genomics, epigenomics, transcriptomics, and reverse genetics. They focused on four species from two distinct genera and showed that differences in the regulation and functionality of a set of homologous MADS-box transcription factors underly the perennial-to-annual transition. Remarkably, transforming Arabidopsis thaliana and Brassica rapa, both annual plants, with the perennial homologs under control of their native promoters led to a perennial-like phenotype. This study provides insights into the molecular mechanisms driving different life strategies in Brassicaceae, emphasizing the role of epigenetic modifications and regulatory genomic regions in the evolution of new traits. (Summary by Carlo Pasini @Crl_Psn) Cell 10.1016/j.cell.2024.04.047
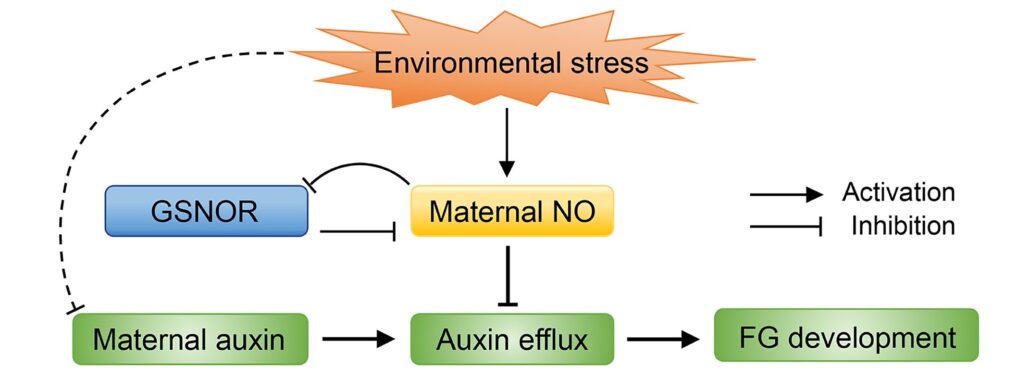
Maternal nitric oxide influences female gametophyte development
Nitric oxide (NO) acts as a signal molecule to regulate plant growth, from seed germination to floral formation. Female gametophyte (FG) development is sensitive to external signals; for example, in plants in adverse environements, a decrease in fertilizable FGs enhances the survival prospects of the offspring. Wang et al. revealed that NO homeostasis in maternal sporophytic (diploid) tissues is essential for FG development in Arabidopsis. Disrupting maternal NO homeostasis leading to elevated NO levels or adding exogenous NO hinders maternal auxin delivery to FGs, impacting FG development. Enhancing maternal control of NO levels or exogenously supplying auxin can boost fertility under stress, suggesting maternal NO homeostasis and auxin supply are key regulators of FG development. The findings point to a mechanism by which plants integrate internal cues and external stress to modulate seed set, with implications for improving crop fertility under adverse conditions. (Summary by Yueh Cho @YuehCho1984) Plant Cell 10.1093/plcell/koae043.
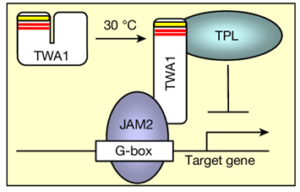
Beat the heat with TWA1, a novel thermosensor in plants
Optimal temperatures for growth and development vary between plant species, making accurate temperature sensing crucial for their fitness. The search for molecular mechanisms underlying temperature signaling suggests that a single temperature sensor likely does not exist in plants. Instead, plants possess a variety of thermosensors, including photosensing-related proteins, RNA thermosensors, and even thermosensitive plasma membranes. In a recent study, Bohn and co-authors identified a novel thermosensor, THERMO-WITH ABA-RESPONSE 1 (TWA1), from a screen for mutants exhibiting ABA hypersensitivity. TWA1 is an intrinsically disordered protein with a highly variable amino-terminal region, which is believed to undergo a temperature-dependent conformational change. This change allows TWA1 nuclear translocation where it interacts with the TOPLESS – JASMONATE-ASSOCIATED MYC-LIKE 2 (TPL-JAM2) co-repressor complex to regulate gene expression and protect plants from heat stress. Interestingly, specific amino acid changes in the amino-terminal region of TWA1 orthologues were linked to different temperature thresholds, related to the climatic conditions where these species grow. Notably, the generation of thermotolerant plants through the overexpression of TWA1 did not negatively impact growth, unlike previously used transgenic strategies that often lead to growth or yield penalties. Therefore, a rational design of a TWA1 gene-editing strategy for generating crops with altered thermal acclimation responses offers a tremendous potential to combat the harmful effects of climate change (Summary by Thomas Depaepe @thdpaepe) Nature 10.1038/s41586-024-07424-x.
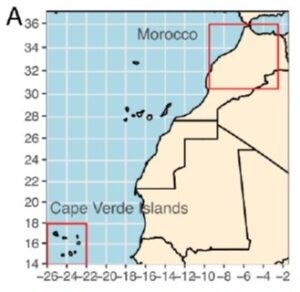
A novel metabolite confers salt tolerance to Arabidopsis from Cape Verde Islands
Rising sea levels and increasing salt accumulation in irrigated soils are threatening food production globally. There is a lot to be learned from plants that have evolved tolerance to salt. In this new preprint, Martínez-Rivas et al. explored the mechanism of salt tolerance the Cvi-0 accession of Arabidopsis that is sourced from the Cape Verde Islands, an island country off the west coast of Africa with very saline soils. Through combined metabolomics and QTL approaches they identified a disaccharide metabolite, which they call U2746, strongly associated with the Cvi-0 allele; this allele also negatively affects levels of glucuronic acid. The causal gene encodes a glycoside hydrolase family 38 protein (GH38cv) previously shown to affect salt tolerance. The authors showed that GH38cv directly controls the accumulation of U2746 and glucuronic acid, with the Cvi-0 allele carrying mutations that affect its function leading to accumulation of U3746. Interestingly, they identified several different mutations in this gene in Arabidopsis plants isolated from the Cape Verde islands, suggesting positive selection for the presence of this disaccharide in these harsh environments. The authors haven’t yet identified what role this novel disaccharide has in salt tolerance (possibly as a signal, osmolyte, or wall modifier), but speculate that it could provide opportunities for agriculture, either through exogenous production or gene editing approaches. (Summary by Mary Williams @PlantTeaching) bioRxiv https://www.biorxiv.org/content/10.1101/2024.05.21.595092v1
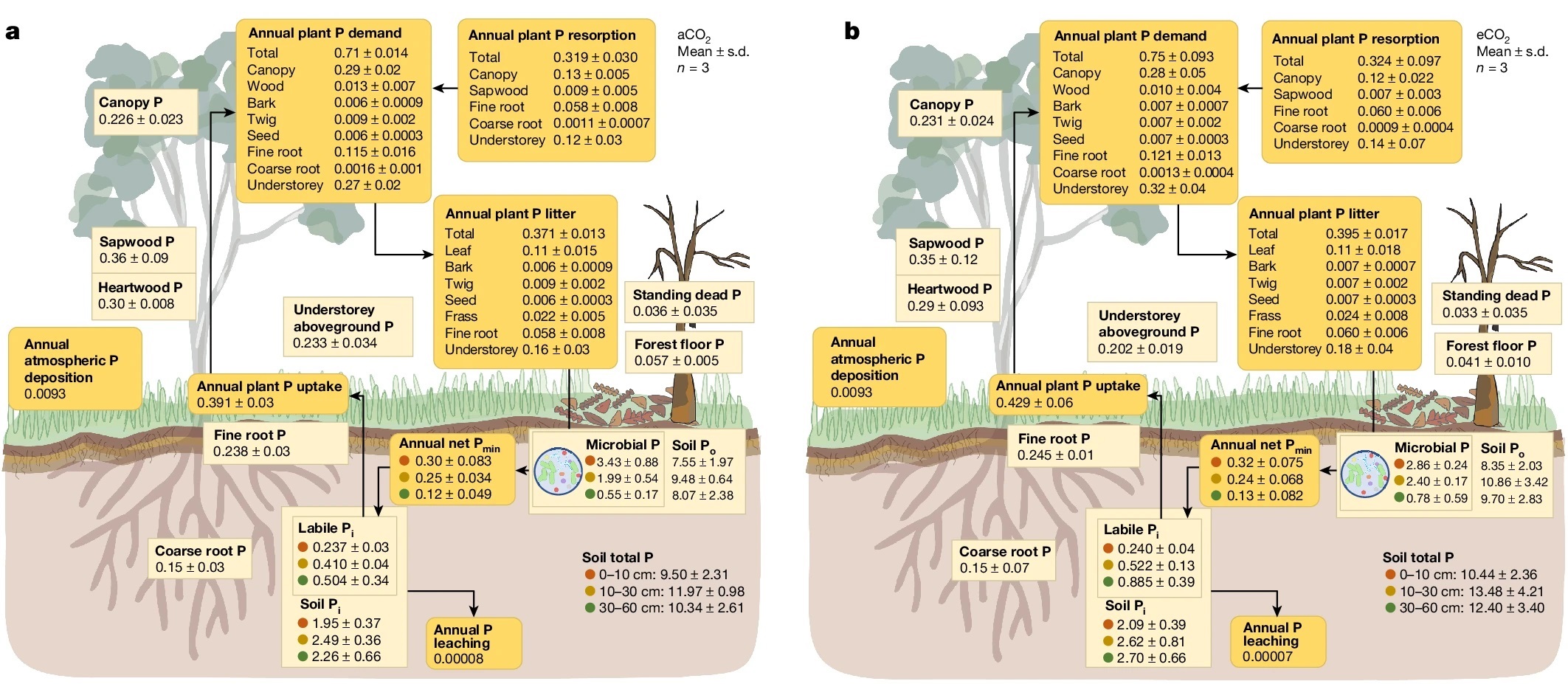
Phosphorus limitation limits carbon storage in CO2-enriched mature forests
Plants fix carbon dioxide to generate biomass, and in many cases increasing the availability of CO2 enhances plant growth. However, some studies have shown that although yields of food crops such as rice might increase, their nutritional value decreases, due to limitations in availability of minerals such as iron and zinc. In this study by Jiang et al., the authors used a Free Air CO2 Enrichment (FACE) system in a eucalyptus forest. Previous studies showed that although CO2 led to increased photosynthesis, this was not reflected in increased biomass. Instead, it led to an increase in root exudation of carbon compounds. The authors speculated that this response might serve as a recruitment signal to soil microbes to aid in plant phosphorus (P) uptake. They undertook a thorough analysis of ecosystem-scale P budget for a mature eucalyptus forest growing in soils with low P-availability (typical of Australian soils). The authors found that: a majority of P in the soil system was present in microbes, enhanced CO2 had little impact on P availability or soil microbe composition, and the competitive superiority of soil microbes with respect to P uptake limits tree growth response to elevated CO2. These findings will be useful in Earth-system models of elevated CO2. (Summary by Mary Williams @PlantTeaching) Nature 10.1038/s41586-024-07491-0
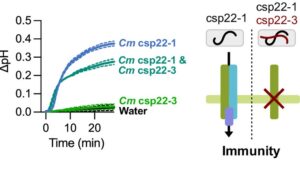
Non-immunogenic bacterial epitopes mask recognition of their immunogenic counterparts
Host plants recognize diverse bacterial epitopes, known as microbe-associated molecular patterns (MAMPs), and respond with an immune reaction to control bacterial growth. However, most studies have focused on single bacterial epitopes, limiting our understanding of plant-bacteria interaction outcomes in real-world scenarios. Stevens et al. took an evolutionary approach to study five common MAMPs in 4,228 plant-associated bacteria. Their analysis showed that these MAMPs are evolutionarily constrained, despite variations in copy numbers (CNVs) and sequences, allowing for experimental testing of host immune responses. Among the MAMPs studied, cold shock proteins (CSPs) were highly diverse, both in CNVs and sequence divergence, and induced a range of immune responses in host plants, from non-immunogenic to highly immunogenic. Further experiments with a Clavibacter strain containing both immunogenic and non-immunogenic CSPs revealed that the non-immunogenic CSPs suppressed the activity of the immunogenic ones within the same strain. Additionally, cell lysates from this strain masked immune induction by other strains, including Streptomyces and the pathogenic Pseudomonas. This comprehensive resource on epitope variations and new insights into how epitopes interact to induce host immunity will help develop strategies to predictably manipulate host-pathogen interactions. (Summary by Arijit Mukherjee @ArijitM61745830) PNAS 10.1073/pnas.2319499121
Leaf yellowing phenotype in rice, mediated by phytoplasma-secreted effector protein, attracts insect vectorsPhytoplasmas are bacterial pathogens that induce significant morphological changes in a host plant including prominent leaf yellowing. They are known to be transmitted by piercing-sucking insects or phloem-feeding arthropods. Phytoplasmas alter plant developmental process through specific effectors. In a study by Zhang et al., an effector protein secreted by rice orange leaf phytoplasma (ROLP) induced leaf yellowing in rice plants. This effector protein, secreted ROLP protein 1 (SRP1), is secreted from ROLP in the infected rice phloem region and translocates to adjacent chloroplasts where it directly targets and negatively regulates the expression of rice glutamine synthetase (OsG2). OsG2 is essential for biosynthesis of chlorophyll precursors, so its impairment by SRP1 causes a leaf yellowing phenotype. Insect two-choice assays revealed that SRP1 overexpression in rice transgenic plants promotes attractiveness for leafhopper pests that transmit ROLP and facilitates ROLP infection into rice phloem. By attracting insect vectors, leaf yellowing is beneficial for ROLP transmission. This is the first study to demonstrate phytoplasma-induced leaf color change attracts arthropod vectors in rice. (Summary by Indrani Kakati @Indranik333) PNAS 10.1073/pnas.2402911121