Plant Science Research Weekly: July 17, 2020
Review: Emerging mechanisms to fine-tune receptor kinase signaling specificity
 Maybe it’s because I’ve spent too much time in lockdown, but sometimes I find studies on cell signaling a bit impenetrable. Fortunately, this excellent review by Galindo-Trigo et al. has arrived, which elegantly walks the reader through the crowded world of receptor-like kinases and “how cells can exert highly localized control over the assembly, interaction and composition of such complexes in order to attain essential cellular output specificity.” As the authors point out, the fact that many signaling components function in multiple diverse pathways provides opportunities for coordination between these pathways (by acting as a node), but also makes it hard to comprehend how signals are properly deciphered. The review takes as an example FERONIA, which participates in many diverse signal and response pathways; its extracellular domain interact with peptides, cell wall components and extensins, and it forms complexes with other RLKs, small LLG proteins, and immune receptors. Specificity of signaling is conferred by small structural variations and the use of different phosphorylation sites, but also dynamic protein trafficking, which includes endocystosis to remove RLKs from the cell surface and also the formation of signaling clusters within membranes. Reading this review brought me one step closer to grasping the nature of these complex and busy but incredibly well orchestrated signaling networks, and the illustrations are excellent. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.05.010
Maybe it’s because I’ve spent too much time in lockdown, but sometimes I find studies on cell signaling a bit impenetrable. Fortunately, this excellent review by Galindo-Trigo et al. has arrived, which elegantly walks the reader through the crowded world of receptor-like kinases and “how cells can exert highly localized control over the assembly, interaction and composition of such complexes in order to attain essential cellular output specificity.” As the authors point out, the fact that many signaling components function in multiple diverse pathways provides opportunities for coordination between these pathways (by acting as a node), but also makes it hard to comprehend how signals are properly deciphered. The review takes as an example FERONIA, which participates in many diverse signal and response pathways; its extracellular domain interact with peptides, cell wall components and extensins, and it forms complexes with other RLKs, small LLG proteins, and immune receptors. Specificity of signaling is conferred by small structural variations and the use of different phosphorylation sites, but also dynamic protein trafficking, which includes endocystosis to remove RLKs from the cell surface and also the formation of signaling clusters within membranes. Reading this review brought me one step closer to grasping the nature of these complex and busy but incredibly well orchestrated signaling networks, and the illustrations are excellent. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.05.010
Modifying plant photosynthesis and growth via simultaneous chloroplast transformation of Rubisco large and small subunits
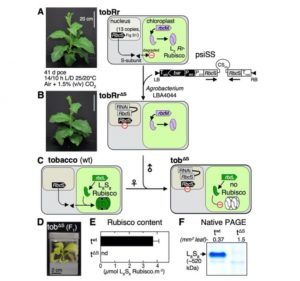 In an interesting evolutionary artifact, the genes encoding the small subunit of Rubisco, rbcS, reside in the nuclear genome, whilst those encoding RbcL persists in the chloroplast. The RbcS protein is translocated into the chloroplast where the holoenzyme forms. This complexity adds to the challenge of engineering Rubsico, and several efforts have been made to overcome it. Here, Martin-Avila et al. have developed a tobacco line with all the endogenous rbcS genes silenced by RNAi, and with the rbcL gene substituted with a bacterial gene that encode the L2 form of Rubisco that does not assemble with the small subunit. These plants can be used to explore new variations of Rubisco. The authors introduced combinations of rbcS and rbcL genes from various sources into the plastid genome and also optimized the rbcS gene for expression and translation in the plastid. These studies allow the activity of engineered Rubisco subunits to be assayed in vivo. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1105/tpc.20.00288
In an interesting evolutionary artifact, the genes encoding the small subunit of Rubisco, rbcS, reside in the nuclear genome, whilst those encoding RbcL persists in the chloroplast. The RbcS protein is translocated into the chloroplast where the holoenzyme forms. This complexity adds to the challenge of engineering Rubsico, and several efforts have been made to overcome it. Here, Martin-Avila et al. have developed a tobacco line with all the endogenous rbcS genes silenced by RNAi, and with the rbcL gene substituted with a bacterial gene that encode the L2 form of Rubisco that does not assemble with the small subunit. These plants can be used to explore new variations of Rubisco. The authors introduced combinations of rbcS and rbcL genes from various sources into the plastid genome and also optimized the rbcS gene for expression and translation in the plastid. These studies allow the activity of engineered Rubisco subunits to be assayed in vivo. (Summary by Mary Williams @PlantTeaching) Plant Cell 10.1105/tpc.20.00288
Variation in maize chlorophyll biosynthesis alters plant architecture
 This is an interesting paper that combines classical genetics with biochemistry and phenotypic analysis. Khangura et al. looked at maize lines with various allelic combinations of two loci that affect chlorophyll biosynthesis: the oil yellow1 (oy1) gene encodes magnesium chelatase, the first committed step in chlorophyll production, and very oil yellow1 (vey1) is a cis-regulatory modifier. Because magnesium chelatase is a multimeric enzyme, some mutant alleles such as Oy1-N1989 behave in a semi-dominant manner. The vey1 modifier allele is a polymorphism of the oy gene that affects mRNA levels, but only is visible phenotypically in plants carrying the Oy1-N1989 allele. The authors found that these allelic interactions can lead to variations in the chlorophyll content of plants, and that the growth responses of the plants are affected by chlorophyll content. Some of these effects are straightforward (e.g., a decrease in stalk width and decreased branching are associated with less chlorophyll) and some less so (e.g., plant height). Double mutant analysis with various genes affecting branching patterns suggests that the chlorophyll concentration acts upstream of branch outgrowth, and the accompanying branching defects are presumably a consequence of decreased energy availability. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00306
This is an interesting paper that combines classical genetics with biochemistry and phenotypic analysis. Khangura et al. looked at maize lines with various allelic combinations of two loci that affect chlorophyll biosynthesis: the oil yellow1 (oy1) gene encodes magnesium chelatase, the first committed step in chlorophyll production, and very oil yellow1 (vey1) is a cis-regulatory modifier. Because magnesium chelatase is a multimeric enzyme, some mutant alleles such as Oy1-N1989 behave in a semi-dominant manner. The vey1 modifier allele is a polymorphism of the oy gene that affects mRNA levels, but only is visible phenotypically in plants carrying the Oy1-N1989 allele. The authors found that these allelic interactions can lead to variations in the chlorophyll content of plants, and that the growth responses of the plants are affected by chlorophyll content. Some of these effects are straightforward (e.g., a decrease in stalk width and decreased branching are associated with less chlorophyll) and some less so (e.g., plant height). Double mutant analysis with various genes affecting branching patterns suggests that the chlorophyll concentration acts upstream of branch outgrowth, and the accompanying branching defects are presumably a consequence of decreased energy availability. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00306
Conserved gene regulatory network integrated with stress response factors in radish and Arabidopsis root cambium
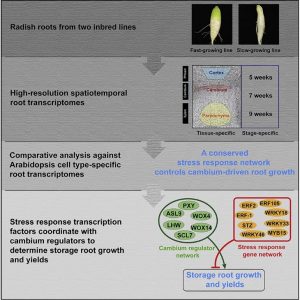 The cambium is a layer of actively dividing cells between xylem and phloem tissues that is responsible for the thickening of primary or lateral roots and stems. Root crops are tightly associated with the cambium regulatory mechanism which is less characterized in root as compared to shoot development. Hoang et al. generated the first high-resolution tissue specific transcriptome analysis from two inbred lines of radish (Raphanus sativus) storage roots, which they compared to their close relative Arabidopsis to understand the co-expression gene-regulatory networks (GRNs). The comparative analysis shows some of the genes in hormone and stress-response pathways are key regulators of the vascular tissue development. This comparative transcriptome analysis further identified a conserved stress-response GRN which is associated with some of the transcription factors that are highly expressed in root cambium. Interestingly, among the transcription factors related to the stress-response root secondary development, the ETHYLENE RESPONSE FACTOR1 (ERF-1) is the key regulator in the network. To test the in vivo cambium GRNs, the authors utilize the gain- and loss-of function alleles to validate ERF-1 as key regulator in the conserved GRNs. Overall, this study provides an insight to fine-tune development in a stressful environment by understanding the network. (Summary by Min May Wong @wongminmay) Curr. Biol. 10.1016/j.cub.2020.05.046
The cambium is a layer of actively dividing cells between xylem and phloem tissues that is responsible for the thickening of primary or lateral roots and stems. Root crops are tightly associated with the cambium regulatory mechanism which is less characterized in root as compared to shoot development. Hoang et al. generated the first high-resolution tissue specific transcriptome analysis from two inbred lines of radish (Raphanus sativus) storage roots, which they compared to their close relative Arabidopsis to understand the co-expression gene-regulatory networks (GRNs). The comparative analysis shows some of the genes in hormone and stress-response pathways are key regulators of the vascular tissue development. This comparative transcriptome analysis further identified a conserved stress-response GRN which is associated with some of the transcription factors that are highly expressed in root cambium. Interestingly, among the transcription factors related to the stress-response root secondary development, the ETHYLENE RESPONSE FACTOR1 (ERF-1) is the key regulator in the network. To test the in vivo cambium GRNs, the authors utilize the gain- and loss-of function alleles to validate ERF-1 as key regulator in the conserved GRNs. Overall, this study provides an insight to fine-tune development in a stressful environment by understanding the network. (Summary by Min May Wong @wongminmay) Curr. Biol. 10.1016/j.cub.2020.05.046
Rocks in the auxin stream: Wound-induced auxin accumulation and ERF115 expression synergistically drive stem cell regeneration
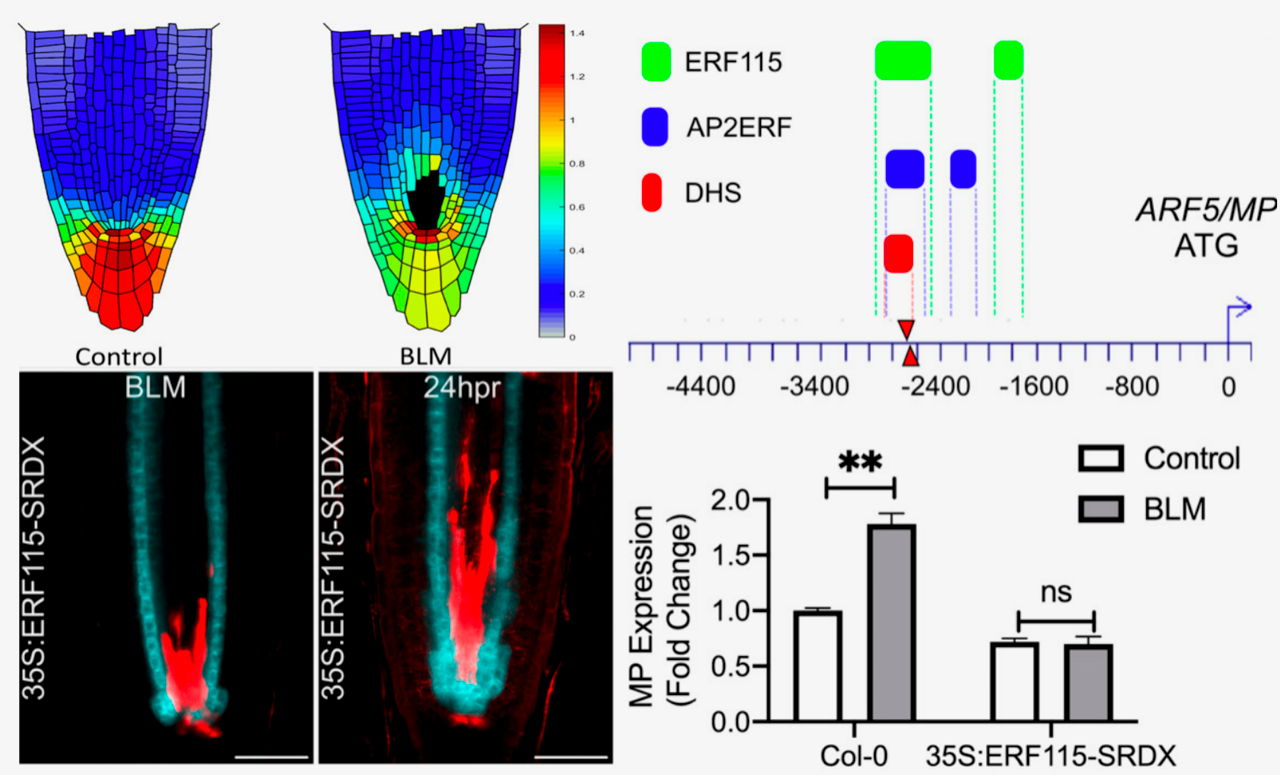 Plants have evolved exquisite regenerative capacities to repair wounds or even reform complete organs thanks to the proliferative activity of stem cell niches residing in meristems. Different types of injuries elicit different developmental and regenerative responses. However, a key player seems to be the phytohormone auxin, which plays multiple roles in wound recovery or regeneration of lost organs. Nevertheless, a mechanistic view of how plants deploy the correct regenerative program against different types of injury is still lacking. A recent paper by Canher et al. sought to investigate cellular regeneration after DNA damage-induced cell death of vascular stem cells. The authors combined in silico modeling of auxin distribution, imaging of reporter, dominant-negative, overexpression and auxin response lines, and treatment with the DNA damage-inducing drug bleomycin (BLM). In their model, dead vascular stem cells obstruct auxin flow (like rocks in a stream) and cause it to accumulate. This activates ERF115 expression, which grants partial stem cell identity to neighboring endodermal cells and fits them with the formative capacity to replace the dead stem cells. RNA-seq analyses of BLM-induced genes showed that ERF115 probably sensitizes cells to auxin by activating expression of ARF5/MONOPTEROS, a major regulator of vascular tissue development, suggesting a mechanism in which ERF115 feeds the wounding input into auxin-dependent developmental processes. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2006620117
Plants have evolved exquisite regenerative capacities to repair wounds or even reform complete organs thanks to the proliferative activity of stem cell niches residing in meristems. Different types of injuries elicit different developmental and regenerative responses. However, a key player seems to be the phytohormone auxin, which plays multiple roles in wound recovery or regeneration of lost organs. Nevertheless, a mechanistic view of how plants deploy the correct regenerative program against different types of injury is still lacking. A recent paper by Canher et al. sought to investigate cellular regeneration after DNA damage-induced cell death of vascular stem cells. The authors combined in silico modeling of auxin distribution, imaging of reporter, dominant-negative, overexpression and auxin response lines, and treatment with the DNA damage-inducing drug bleomycin (BLM). In their model, dead vascular stem cells obstruct auxin flow (like rocks in a stream) and cause it to accumulate. This activates ERF115 expression, which grants partial stem cell identity to neighboring endodermal cells and fits them with the formative capacity to replace the dead stem cells. RNA-seq analyses of BLM-induced genes showed that ERF115 probably sensitizes cells to auxin by activating expression of ARF5/MONOPTEROS, a major regulator of vascular tissue development, suggesting a mechanism in which ERF115 feeds the wounding input into auxin-dependent developmental processes. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2006620117
Seed traits determine species responses to fire under varying soil heating scenarios ($)
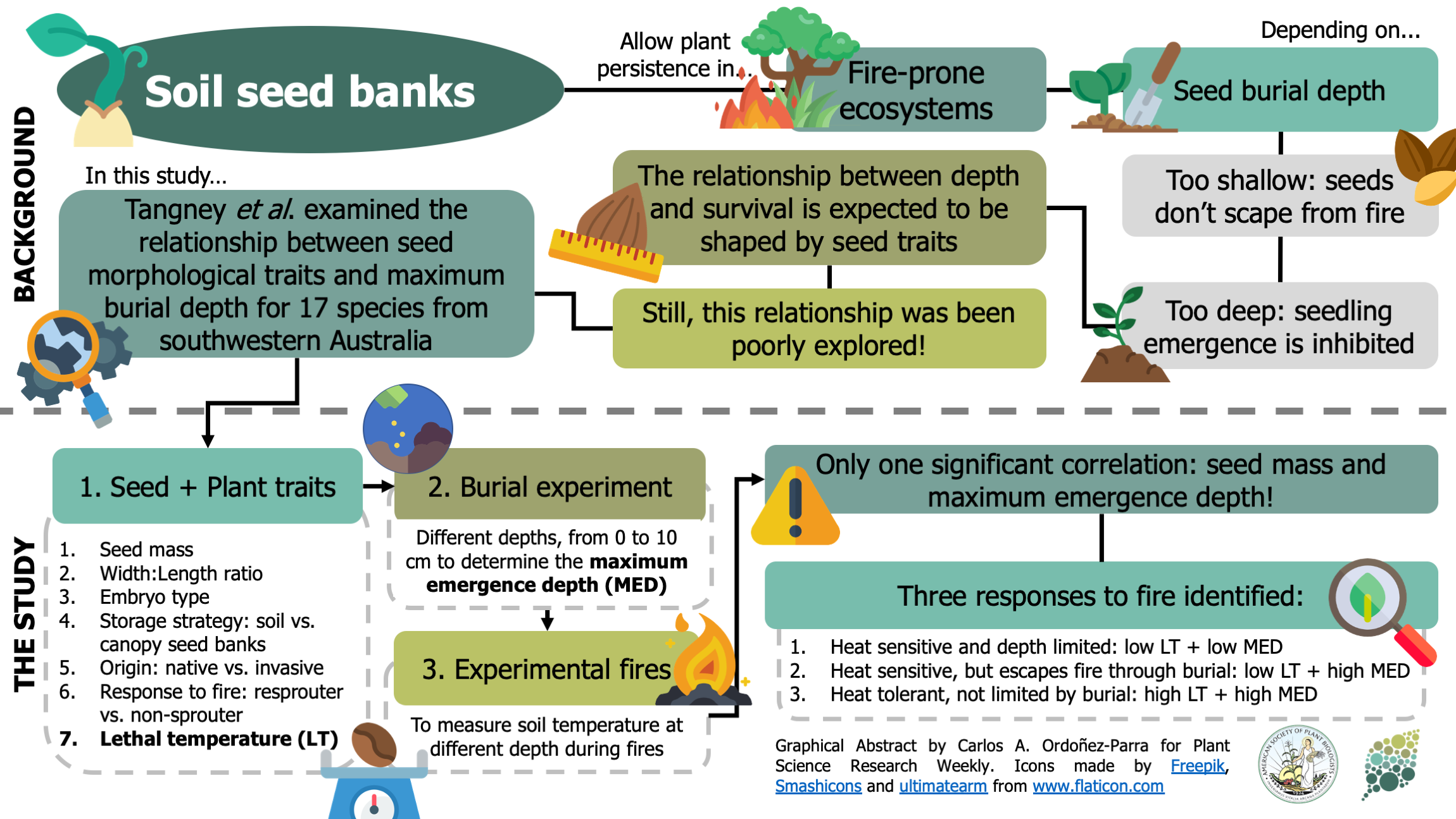 Soil seed banks allow plants to persist in fire-prone ecosystems. However, if seeds are buried too deep, seedling emergence can be inhibited. The maximum depth at which seeds can be buried without hampering recruitment is expected to be shaped by different seed traits, although this has been seldom tested. Here, Tangney et al. examined the relationship between seed traits and maximum burial depth. Through a combination of greenhouse and laboratory experiments, the authors assessed the lethal temperature (i.e., the temperature where 50% of seeds lose viability) and maximum emergence depth of 17 species from southwestern Australia. Experimental fires of different intensities were also conducted to measure soil temperatures at different depths. Seed mass was positively correlated with maximum emergence depth, suggesting heavier seeds had more abundant reserves that allowed them to emerge from deeper in the soil. Considering the soil temperatures registered during experimental fires, the authors describe a spectrum in fire sensitivity: from highly sensitive species (low lethal temperatures and maximum emergence depth) to highly resistant species (higher lethal temperatures an emergence depths). As a result, this research provides interesting insights for future efforts about the role of functional traits in plant regeneration in fire-prone ecosystems. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Funct. Ecol. 10.1111/1365-2435.13623
Soil seed banks allow plants to persist in fire-prone ecosystems. However, if seeds are buried too deep, seedling emergence can be inhibited. The maximum depth at which seeds can be buried without hampering recruitment is expected to be shaped by different seed traits, although this has been seldom tested. Here, Tangney et al. examined the relationship between seed traits and maximum burial depth. Through a combination of greenhouse and laboratory experiments, the authors assessed the lethal temperature (i.e., the temperature where 50% of seeds lose viability) and maximum emergence depth of 17 species from southwestern Australia. Experimental fires of different intensities were also conducted to measure soil temperatures at different depths. Seed mass was positively correlated with maximum emergence depth, suggesting heavier seeds had more abundant reserves that allowed them to emerge from deeper in the soil. Considering the soil temperatures registered during experimental fires, the authors describe a spectrum in fire sensitivity: from highly sensitive species (low lethal temperatures and maximum emergence depth) to highly resistant species (higher lethal temperatures an emergence depths). As a result, this research provides interesting insights for future efforts about the role of functional traits in plant regeneration in fire-prone ecosystems. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Funct. Ecol. 10.1111/1365-2435.13623
Extracellular proteolytic cascade in tomato activates immune protease Rcr3
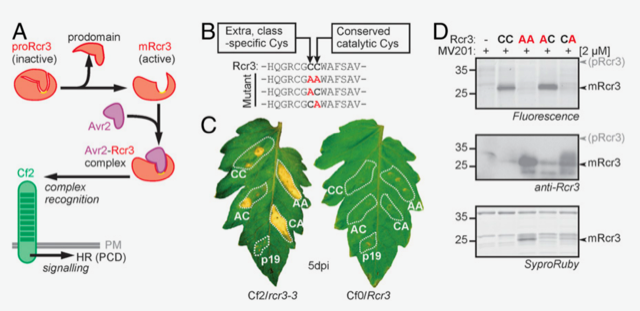 The plant apoplast is a sea of immune-related proteins that facilitate robust defense against pathogens. Rcr3 is a tomato secreted apoplastic protease that contributes to both basal defense and gene-for-gene resistance against pathogens. Rcr3 is activated by the cleavage of its autoinhibitory prodomain, and it has been considered that the catalytic activity of Rcr3 is required for its maturation. Paulus et al. however, found that catalytically inactive Rcr3 can be matured and functional in plants. The authors screened for other proteases that process Rcr3 and identified a subtilisin (SBT), P69B, from tomato apoplastic fluid. P69B cleaves Asp residues of Rcr3 in vitro and is required for Rcr3 processing in tomato plants. The maturation of Rcr3 is dispensable for gene-for-gene resistance processes but potentially required for basal defense against pathogens. Interestingly, the oomycete pathogen P. infestans secrets effector proteins that inhibit multiple SBTs, possibly as a strategy to overcome Rcr3-mediated plant immunity. Furthermore, a distantly related SBT in N. benthamiana, and probably other SBTs, can also process Rcr3, suggesting that the activation of Rcr3 by SBT may be widespread across the plant kingdom. This study demonstrates the first proteolytic cascade in the plant kingdom, illuminating robust apoplastic immune signaling. (Summary by Tatsuya Nobori @nobolly) PNAS 10.1073/pnas.1921101117
The plant apoplast is a sea of immune-related proteins that facilitate robust defense against pathogens. Rcr3 is a tomato secreted apoplastic protease that contributes to both basal defense and gene-for-gene resistance against pathogens. Rcr3 is activated by the cleavage of its autoinhibitory prodomain, and it has been considered that the catalytic activity of Rcr3 is required for its maturation. Paulus et al. however, found that catalytically inactive Rcr3 can be matured and functional in plants. The authors screened for other proteases that process Rcr3 and identified a subtilisin (SBT), P69B, from tomato apoplastic fluid. P69B cleaves Asp residues of Rcr3 in vitro and is required for Rcr3 processing in tomato plants. The maturation of Rcr3 is dispensable for gene-for-gene resistance processes but potentially required for basal defense against pathogens. Interestingly, the oomycete pathogen P. infestans secrets effector proteins that inhibit multiple SBTs, possibly as a strategy to overcome Rcr3-mediated plant immunity. Furthermore, a distantly related SBT in N. benthamiana, and probably other SBTs, can also process Rcr3, suggesting that the activation of Rcr3 by SBT may be widespread across the plant kingdom. This study demonstrates the first proteolytic cascade in the plant kingdom, illuminating robust apoplastic immune signaling. (Summary by Tatsuya Nobori @nobolly) PNAS 10.1073/pnas.1921101117
Optimizing PollenCounter for high throughput phenotyping of pollen quality in tomatoes
 PollenCounter is an open source, ImageJ based macro that splits RGB images of stained pollen grains into the primary channels. The program estimates pollen viability in plants through analyzing red (total number of pollen grains) and green (only viable pollen grains) images, after which the particles are counted. This macro has been used to assess pollen viability in grapevine, however a method for optimizing it have not been developed in tomatoes. Here, Ayenan et al. developed an optimization method for assessing pollen quality in tomatoes. First, they counted tomato pollen grains manually and determined viability. Next, they used the original macro parameters developed for grape (pollen size of 60–800 pixel2 and circularity of 0.4-1). This process indicated over 100% viability demonstrating that the original paramters overestimated the number of viable pollens in tomatoes. To correct this, the authors used six optimization scenarios (ranging from a circularity of 0.4–1 to 0.6-1 and pollen size of 100–900 to 200-900 pixels2) to adjust the original macro. At a circularity of 0.4-1 and pollen size of 100–900 pixel2, they got the highest correlation for pollen number and viability as assessed manually. Furthermore, they used this optimized PollenCounter to discriminate between genotypes of heat stressed tomatoes and recommended best practices to improve the accuracy of a PollenCounter. The use of PollenCounter will save time and money and improve the efficacy of pollen quality assessment in breeding programs. (Summary by Modesta N. Abugu @modestannedi). MethodsX. 10.1016/j.mex.2020.100977
PollenCounter is an open source, ImageJ based macro that splits RGB images of stained pollen grains into the primary channels. The program estimates pollen viability in plants through analyzing red (total number of pollen grains) and green (only viable pollen grains) images, after which the particles are counted. This macro has been used to assess pollen viability in grapevine, however a method for optimizing it have not been developed in tomatoes. Here, Ayenan et al. developed an optimization method for assessing pollen quality in tomatoes. First, they counted tomato pollen grains manually and determined viability. Next, they used the original macro parameters developed for grape (pollen size of 60–800 pixel2 and circularity of 0.4-1). This process indicated over 100% viability demonstrating that the original paramters overestimated the number of viable pollens in tomatoes. To correct this, the authors used six optimization scenarios (ranging from a circularity of 0.4–1 to 0.6-1 and pollen size of 100–900 to 200-900 pixels2) to adjust the original macro. At a circularity of 0.4-1 and pollen size of 100–900 pixel2, they got the highest correlation for pollen number and viability as assessed manually. Furthermore, they used this optimized PollenCounter to discriminate between genotypes of heat stressed tomatoes and recommended best practices to improve the accuracy of a PollenCounter. The use of PollenCounter will save time and money and improve the efficacy of pollen quality assessment in breeding programs. (Summary by Modesta N. Abugu @modestannedi). MethodsX. 10.1016/j.mex.2020.100977



