Plant Science Research Weekly: July 10, 2020
Review: Banishing barberry: The history of Berberis vulgaris prevalence and wheat stem rust incidence across Britain

Puccinia graminis f. sp. tritici (Pgt) has a strange life strategy in that it requires two hosts to complete its sexual life cycle; wheat and barberry. In the absence of barberry, it reproduces clonally; clonal reproduction limits its ability to generate diversity and therefore its ability to evade plant defenses. Barnes et al. take a historical look at how people in Britain have viewed barberry, starting from the gradual awakening that it does in fact promote the spread of this critically dangerous pathogen, which led to campaigns to remove it. This history is particularly relevant today because some have argued that, as barberry is the sole food source for the endangered barberry carpet moth (Pareulype berberata), it should be cultivated in Britain in spite of its contribution to the spread of a disease that impacts a hugely important crop. What course of action does history support? (You’ll have to look at the article to find out!). (Summary by Mary Williams @PlantTeaching) Plant Pathology. 10.1111/ppa.13231
Pan-genome of wild and cultivated soybeans
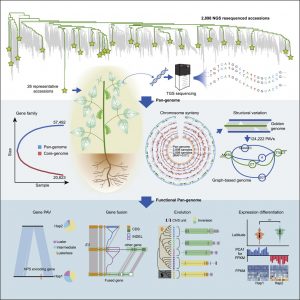 Widespread genome sequencing of individuals has revealed the high level of intraspecific variability in plant species. As such, constructing high-quality pan-genomes is a growing necessity to study dynamic plant genomes. Here, Liu et al. release the soybean pan-genome formed from sequencing 2,898 accessions, including landraces, wild, and cultivated soybeans. The authors de novo assembled 26 accessions which were carefully selected to represent the geographical distributions and phylogenetics of the 2,898 accessions. A graph-based genome was subsequently formed, onto which short reads of the 2,898 accessions were mapped, identifying over 55,000 structural variants (SVs). The authors found 15 gene fusion events in different accessions, including a gene fusion involving E3, a PHYA ortholog. Furthermore, the contribution of SVs in soybean domestication was highlighted, such as the alteration of seed coat pigmentation. Finally, over 1,000 SVs in core genes (present in all newly assembled accessions) were associated with altered expression levels, and an iron deficiency chlorosis QTL was clarified, with SVs at this locus uncovered in various accessions. The de novo genomes assembled in this study are promising candidates for sophisticated soybean mapping population construction given that they were carefully selected to encompass both phylogenetic and geographic distributions. The publication of this high-quality pan-genome signifies a comprehensive resource that will certainly expedite both future breeding efforts and evolutionary studies in soybean. (Summary by Caroline Dowling @CarolineD0wling) Cell 10.1016/j.cell.2020.05.023
Widespread genome sequencing of individuals has revealed the high level of intraspecific variability in plant species. As such, constructing high-quality pan-genomes is a growing necessity to study dynamic plant genomes. Here, Liu et al. release the soybean pan-genome formed from sequencing 2,898 accessions, including landraces, wild, and cultivated soybeans. The authors de novo assembled 26 accessions which were carefully selected to represent the geographical distributions and phylogenetics of the 2,898 accessions. A graph-based genome was subsequently formed, onto which short reads of the 2,898 accessions were mapped, identifying over 55,000 structural variants (SVs). The authors found 15 gene fusion events in different accessions, including a gene fusion involving E3, a PHYA ortholog. Furthermore, the contribution of SVs in soybean domestication was highlighted, such as the alteration of seed coat pigmentation. Finally, over 1,000 SVs in core genes (present in all newly assembled accessions) were associated with altered expression levels, and an iron deficiency chlorosis QTL was clarified, with SVs at this locus uncovered in various accessions. The de novo genomes assembled in this study are promising candidates for sophisticated soybean mapping population construction given that they were carefully selected to encompass both phylogenetic and geographic distributions. The publication of this high-quality pan-genome signifies a comprehensive resource that will certainly expedite both future breeding efforts and evolutionary studies in soybean. (Summary by Caroline Dowling @CarolineD0wling) Cell 10.1016/j.cell.2020.05.023
An inducible genome editing system for plants
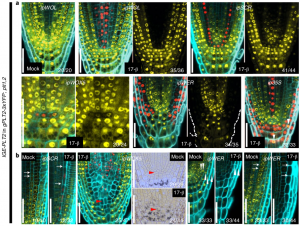 To understand gene function, it is necessary to study loss-of-function phenotypes. However, knockouts of some genes are lethal, which makes it impossible to study their loss-of-function phenotype. Although here are inducible overexpression systems and inducible knockdown systems, as yet there has been no inducible knockout system. In this paper Wang et al. developed an inducible genome editing (IGE) system by a two-step gateway cloning for conditional gene knockout. This IGE depends on the three components brought together in a plant binary vector, 1) a chimeric protein called XVE, that can be activated by estradiol and binds to the LexA binding sites in the promoter to activate the expression of the downstream gene, 2) Cas9 expressed under LexA promoter, 3) sgRNA(s) targeting the desired gene. IGE can be used in a tissue-specific or cell type-specific manner by cloning XVE under tissue-specific or cell type-specific promoters. In this paper the authors examined the effects of knocking out PLETHORA, which encodes an AP2/EREBP transcription factor, RETINOBLASTOMA-RELATED (RBR), which encodes a cell cycle regulator, and GNOM, which encode a brefeldin A sensitive ARF guanine-nucleotide exchange factor. The authors confirmed their conditional knockouts in both a broad domain and cell type-specific domain, and they compared IGE with inducible artificial microRNAs and showed that IGE is more specific. In a nutshell, this paper provides a system for efficient, precise inducible knockout of genes in Arabidopsis. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/s41477-020-0695-2
To understand gene function, it is necessary to study loss-of-function phenotypes. However, knockouts of some genes are lethal, which makes it impossible to study their loss-of-function phenotype. Although here are inducible overexpression systems and inducible knockdown systems, as yet there has been no inducible knockout system. In this paper Wang et al. developed an inducible genome editing (IGE) system by a two-step gateway cloning for conditional gene knockout. This IGE depends on the three components brought together in a plant binary vector, 1) a chimeric protein called XVE, that can be activated by estradiol and binds to the LexA binding sites in the promoter to activate the expression of the downstream gene, 2) Cas9 expressed under LexA promoter, 3) sgRNA(s) targeting the desired gene. IGE can be used in a tissue-specific or cell type-specific manner by cloning XVE under tissue-specific or cell type-specific promoters. In this paper the authors examined the effects of knocking out PLETHORA, which encodes an AP2/EREBP transcription factor, RETINOBLASTOMA-RELATED (RBR), which encodes a cell cycle regulator, and GNOM, which encode a brefeldin A sensitive ARF guanine-nucleotide exchange factor. The authors confirmed their conditional knockouts in both a broad domain and cell type-specific domain, and they compared IGE with inducible artificial microRNAs and showed that IGE is more specific. In a nutshell, this paper provides a system for efficient, precise inducible knockout of genes in Arabidopsis. (Summary by Vijaya Batthula @Vijaya_Batthula) Nature Plants 10.1038/s41477-020-0695-2
An optogenetic tool for plants grown in ambient light
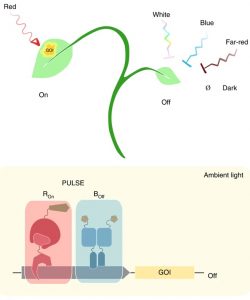 Optogenetics has revolutionized mammalian biology by its ability to fine-tune gene expression in individual cells by pulses of light. However, its application in plants has been challenging because the white light required for plant growth misfires the system. Ochoa-Fernandez et al. solved this problem by engineered photoreceptors that can repress and induce gene expression driven by blue and red light, respectively. The system, termed plant usable light-switch elements (PULSE), allows quantitative and spatiotemporal reversible control of gene expression without being influenced by the broad-spectrum of white light. To guide experimental conditions, the authors developed a quantitative model that predicts PULSE activity. The authors devised two strategies to control gene expression: 1) inducing and targeting a transcriptional activator to the promoter region of the gene of interest using Cas9 and guide RNA and 2) inducing a plant endogenous transcription factor that controls expression of its target genes. They demonstrated that an optogenetically-induced immune receptor could control plant immune responses in Nicotiana benthamiana transiently transformed with PULSE. They also applied this system to conditionally localize proteins to a specific sub-cellular domain of the cell using nanobodies that have a high affinity to particular proteins. Finally, they showed PULSE is functional in stable Arabidopsis transgenic lines. This new tool facilitates the precise manipulation of biological processes in plants to address fundamental questions in plant biology. (Summary by Tatsuya Nobori @nobolly) Nature Methods 10.1038/s41592-020-0868-y
Optogenetics has revolutionized mammalian biology by its ability to fine-tune gene expression in individual cells by pulses of light. However, its application in plants has been challenging because the white light required for plant growth misfires the system. Ochoa-Fernandez et al. solved this problem by engineered photoreceptors that can repress and induce gene expression driven by blue and red light, respectively. The system, termed plant usable light-switch elements (PULSE), allows quantitative and spatiotemporal reversible control of gene expression without being influenced by the broad-spectrum of white light. To guide experimental conditions, the authors developed a quantitative model that predicts PULSE activity. The authors devised two strategies to control gene expression: 1) inducing and targeting a transcriptional activator to the promoter region of the gene of interest using Cas9 and guide RNA and 2) inducing a plant endogenous transcription factor that controls expression of its target genes. They demonstrated that an optogenetically-induced immune receptor could control plant immune responses in Nicotiana benthamiana transiently transformed with PULSE. They also applied this system to conditionally localize proteins to a specific sub-cellular domain of the cell using nanobodies that have a high affinity to particular proteins. Finally, they showed PULSE is functional in stable Arabidopsis transgenic lines. This new tool facilitates the precise manipulation of biological processes in plants to address fundamental questions in plant biology. (Summary by Tatsuya Nobori @nobolly) Nature Methods 10.1038/s41592-020-0868-y
Fluctuating auxin response gradients determine pavement cell-shape acquisition
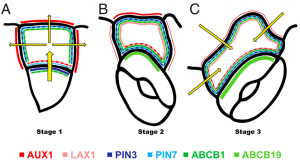 The leaf epidermis is composed primarily of undulated pavement cells arranged in a jigsaw puzzle-like architecture, with neighboring cells flawlessly interlacing with one another thanks to synchronized growth, thus making it an ideal model to study morphogenesis regulation. Seeking to better understand cell-shape acquisition, a recent study by Grones et al. used a subpopulation of leaf epidermal pavement cells known as stomatal lineage ground cells (SLGCs) to address the question of how the formation of lobes in these cells is regulated. By quantifying the outgrowth angle of the first lobe and through the use of an auxin overproduction mutant, the authors determined that lobe outgrowth occurs perpendicular to the stoma (pore) and that this lobe formation is an auxin dose-dependent process. Furthermore, by analyzing auxin response in SLGCs using auxin-reporter DR5 lines the authors showed that auxin response gradients that are fueled by auxin transport, rather than local auxin biosynthesis, regulate lobe formation in SLGCs, and that this gradient shifts dynamically throughout lobe formation due to changes in auxin transporter localization. This paper highlights the importance of dynamic auxin response gradients, how these are controlled by complex auxin transporter localization patterns, and how these processes cooperate to regulate cell shape. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2007400117
The leaf epidermis is composed primarily of undulated pavement cells arranged in a jigsaw puzzle-like architecture, with neighboring cells flawlessly interlacing with one another thanks to synchronized growth, thus making it an ideal model to study morphogenesis regulation. Seeking to better understand cell-shape acquisition, a recent study by Grones et al. used a subpopulation of leaf epidermal pavement cells known as stomatal lineage ground cells (SLGCs) to address the question of how the formation of lobes in these cells is regulated. By quantifying the outgrowth angle of the first lobe and through the use of an auxin overproduction mutant, the authors determined that lobe outgrowth occurs perpendicular to the stoma (pore) and that this lobe formation is an auxin dose-dependent process. Furthermore, by analyzing auxin response in SLGCs using auxin-reporter DR5 lines the authors showed that auxin response gradients that are fueled by auxin transport, rather than local auxin biosynthesis, regulate lobe formation in SLGCs, and that this gradient shifts dynamically throughout lobe formation due to changes in auxin transporter localization. This paper highlights the importance of dynamic auxin response gradients, how these are controlled by complex auxin transporter localization patterns, and how these processes cooperate to regulate cell shape. (Summary by Jesus Leon @jesussaur) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2007400117
A CEP Peptide receptor-like kinase regulates root growth and symbiotic nodulation
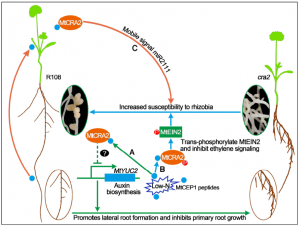 Under low N condition, plants form more lateral roots to facilitate N-uptake efficiency, and legumes such as Medicago truncatula develop more nodules to fulfill plant nitrogen requirements. Since both lateral root formation and nodulation are energy-consuming processes, plants balance these developmental processes, but the regulatory mechanism involved is still a question. Zhu et al. found that MtCEP1-peptide interaction with the MtCRA2 (Compact Root Architecture 2) receptor-like Kinase is involved in coordinating root and nodule development. The authors performed transcriptome analysis of cra2 mutants and showed that under low nitrogen conditions, MtCRA2 inhibits the expression of MtYUC2 (YUCCA2) as well as other auxin transport and metabolism genes thereby modulating the root system architecture. The nodulation phenotype of the ein2 cra2 double mutant and in-vitro phosphorylation assays showed that MtCRA2 can trans-phosphorylate MtEIN2 and inhibit MtEIN2 cleavage, interrupting the ethylene signaling pathway and making plant roots more susceptible to rhizobial infection. The authors propose a model showing the role of the MtCEP1/MtCRA2 pathway in regulating auxin and ethylene hormone signaling pathways to balance lateral root growth and nodulation under low N condition. (Summary by Sunita Pathak @psunita980) Plant Cell 10.1105/tpc.19.00428
Under low N condition, plants form more lateral roots to facilitate N-uptake efficiency, and legumes such as Medicago truncatula develop more nodules to fulfill plant nitrogen requirements. Since both lateral root formation and nodulation are energy-consuming processes, plants balance these developmental processes, but the regulatory mechanism involved is still a question. Zhu et al. found that MtCEP1-peptide interaction with the MtCRA2 (Compact Root Architecture 2) receptor-like Kinase is involved in coordinating root and nodule development. The authors performed transcriptome analysis of cra2 mutants and showed that under low nitrogen conditions, MtCRA2 inhibits the expression of MtYUC2 (YUCCA2) as well as other auxin transport and metabolism genes thereby modulating the root system architecture. The nodulation phenotype of the ein2 cra2 double mutant and in-vitro phosphorylation assays showed that MtCRA2 can trans-phosphorylate MtEIN2 and inhibit MtEIN2 cleavage, interrupting the ethylene signaling pathway and making plant roots more susceptible to rhizobial infection. The authors propose a model showing the role of the MtCEP1/MtCRA2 pathway in regulating auxin and ethylene hormone signaling pathways to balance lateral root growth and nodulation under low N condition. (Summary by Sunita Pathak @psunita980) Plant Cell 10.1105/tpc.19.00428
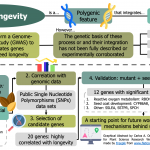
Identification of novel seed longevity genes related to oxidative stress and seed coat by GWAS and reverse genetics
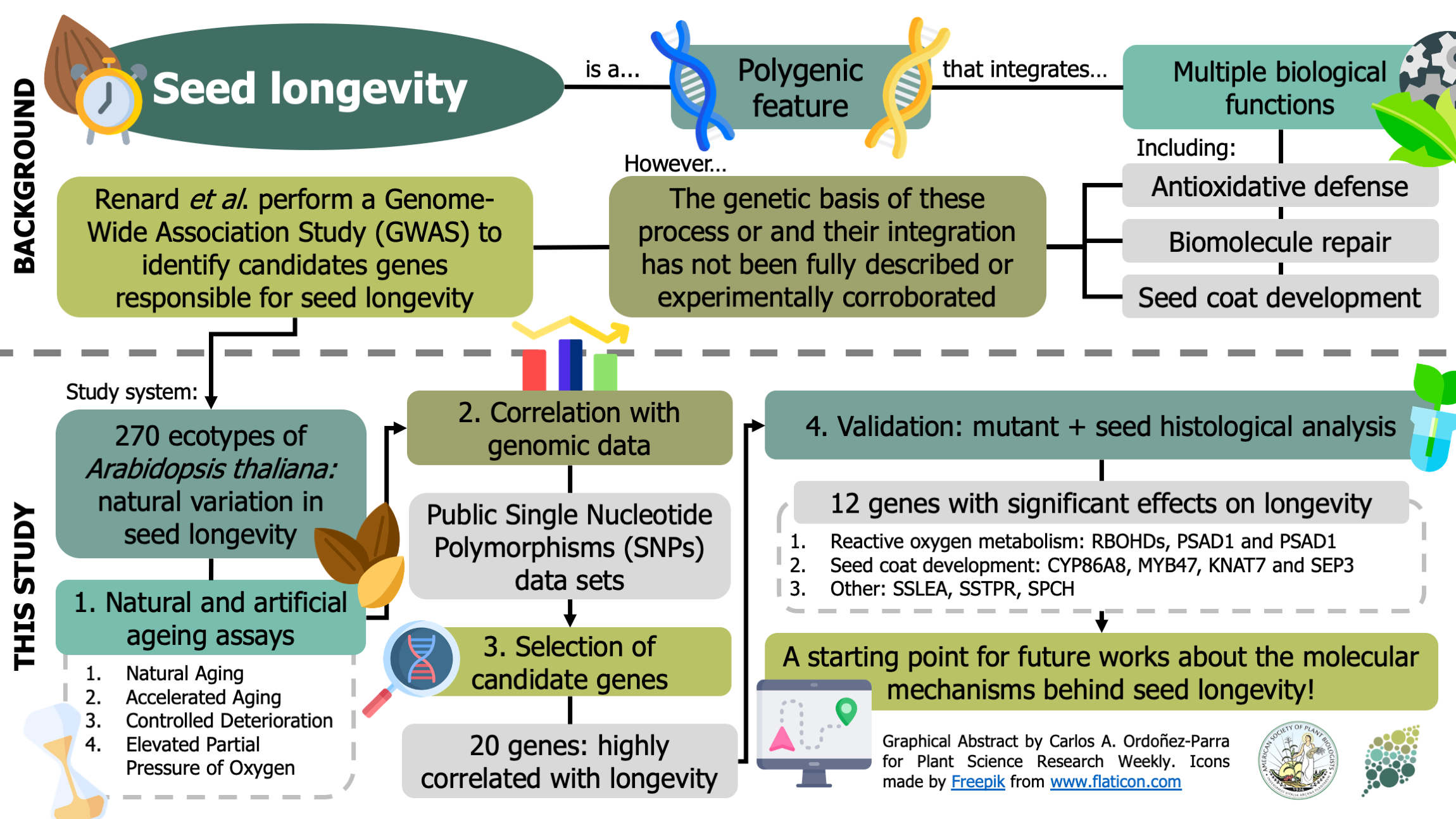
Seed longevity (i.e., the inherent lifespan of a seed) is a polygenic feature that demands the coordination of multiple biological processes, and that shapes the persistence of seeds in the soil and the time they can be stored ex situ. Recent studies on this trait highlight the role of the antioxidant and biomolecule repair systems and the formation of the seed coat. However, the genetic basis behind their contribution has not yet been fully described or corroborated. Here, Renard et al. perform a Genome-Wide Association Study (GWAS) to identify the genes responsible for the differences in the seed longevity of 270 ecotypes of Arabidopsis thaliana. Seeds were exposed to high humidity, high temperature or elevated oxygen pressure conditions (or a combination of these) in order to accelerate the loss of their viability. The results of these assays were correlated with the genomic data available on public databases. After this, 20 candidate genes were validated through a mutant analysis, and 12 had a significant effect in longevity. Most genes were related to enzymes that participate in the metabolism of reactive oxygen species, such as NADPH oxidases (RBOHs) and dehydroascorbate reductases (DHAR1). A cytochrome hydroxylase (CYP86A8) and several transcription factors related to seed coat development also promoted longevity. This paper provides a promising starting point for future research about the mechanisms behind seed longevity. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Cell Environ. 10.1111/pce.13822