Plant Science Research Weekly: January 10, 2025
Focus Issue: Hypoxia and Plants
 The January 2025 issue of Plant Physiology has a focus on “Hypoxia and Plants”. This field has made a lot of progress recently in understanding plant responses to low oxygen, from the molecular to physiological and developmental levels. The focus issue includes reviews on topics such as divergent responses of rice to flooding, how cells perceive and respond to hypoxia, its effects on seeds, tools and synthetic biology approaches to investigate hypoxia, and a very interesting story of the ecology of hypoxia, looking at genetic adaptations in high-altitude plants. The issue also includes research articles on hydraulic and transcriptional responses to water-logging and hypoxia. The editors of the focus issue have invited four speakers whose work appears in the focus issue to speak in a webinar, scheduled for January 28, see https://blog.aspb.org/january-28-plant-physiology-webinar-hypoxia-and-plants/. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Plant Physiol. https://academic.oup.com/plphys/issue#2085436-7695743
The January 2025 issue of Plant Physiology has a focus on “Hypoxia and Plants”. This field has made a lot of progress recently in understanding plant responses to low oxygen, from the molecular to physiological and developmental levels. The focus issue includes reviews on topics such as divergent responses of rice to flooding, how cells perceive and respond to hypoxia, its effects on seeds, tools and synthetic biology approaches to investigate hypoxia, and a very interesting story of the ecology of hypoxia, looking at genetic adaptations in high-altitude plants. The issue also includes research articles on hydraulic and transcriptional responses to water-logging and hypoxia. The editors of the focus issue have invited four speakers whose work appears in the focus issue to speak in a webinar, scheduled for January 28, see https://blog.aspb.org/january-28-plant-physiology-webinar-hypoxia-and-plants/. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Plant Physiol. https://academic.oup.com/plphys/issue#2085436-7695743
Special issue: Parasitic plants
 Runo, Wicke, and Thorogood have edited a special issue of Plants, People, Planet on the topic of parasitic plants. (Note – the Special Issue will be launched on February 19, but the articles are already online in Early View). It’s nice to see a collection of articles that focus on a phylogenetically diverse collection of plants that share an unusual life trait, the ability (or requirement) to parasitize other plants. The collection highlights three major points. First, a few parasitic plants are major threats to food security; these include witchweeds (e.g., Striga spp) and broomrapes (e.g., Orobanche spp), so understanding their biology is of practical importance. Second, a few are endoparasites that live nearly their full life inside of other plants, and some of these are endangered, including Rafflesia spp. that produce the world’s largest flowers. Finally, there are fascinating comparative studies that can be made amongst these very special plants. Key questions include how the cross-species communication that allows parasitic seeds detect a suitable host, triggering germination; how some hosts are resistant and others susceptible; and how the parasite pushes into the host tissues and evades its defenses. There’s a lot of fascinating biology in this issue and much of it is inherently interesting due to the strange nature of parasitic plants. Check the website after the official launch on February 19 for the cover reveal! (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Plants, People, Planet https://doi.org/10.1002/ppp3.10616
Runo, Wicke, and Thorogood have edited a special issue of Plants, People, Planet on the topic of parasitic plants. (Note – the Special Issue will be launched on February 19, but the articles are already online in Early View). It’s nice to see a collection of articles that focus on a phylogenetically diverse collection of plants that share an unusual life trait, the ability (or requirement) to parasitize other plants. The collection highlights three major points. First, a few parasitic plants are major threats to food security; these include witchweeds (e.g., Striga spp) and broomrapes (e.g., Orobanche spp), so understanding their biology is of practical importance. Second, a few are endoparasites that live nearly their full life inside of other plants, and some of these are endangered, including Rafflesia spp. that produce the world’s largest flowers. Finally, there are fascinating comparative studies that can be made amongst these very special plants. Key questions include how the cross-species communication that allows parasitic seeds detect a suitable host, triggering germination; how some hosts are resistant and others susceptible; and how the parasite pushes into the host tissues and evades its defenses. There’s a lot of fascinating biology in this issue and much of it is inherently interesting due to the strange nature of parasitic plants. Check the website after the official launch on February 19 for the cover reveal! (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Plants, People, Planet https://doi.org/10.1002/ppp3.10616
BOOSTER: Unlocking photosynthetic efficiency for enhanced plant productivity
 Photosynthesis is a fundamental biological process for carbon fixation and capturing light energy to drive plant growth. However, excessive sunlight can cause photodamage, which is detrimental to plant health. To protect the photosynthetic machinery from over-excitation and subsequent damage, plant cells employ a sophisticated mechanism to regulate the distribution of excitation energy between the reaction centers and the antenna complexes—a process known as non-photochemical quenching (NPQ). The rapid relaxation of NPQ under low-light conditions and its swift induction in high-light environments are critical for preventing photodamage. However, the slow adjustment of NPQ to fluctuating light is estimated to reduce potential carbon gain by up to 40%. Accelerating the dynamic adjustment of NPQ to changing light conditions could, in principle, significantly enhance net photosynthetic efficiency. Using genome-wide association studies (GWAS), Feyissa and colleagues identified BOOSTER as a putative gene associated with NPQ in poplar. Overexpressing this gene increased photosynthetic efficiency and enhanced biomass production by up to 200% in both poplar and Arabidopsis. This discovery offers a promising strategy to improve plant productivity, with broad potential applications across various plant species. (Summary by Ching Chan @ntnuchanlab) Developmental Cell 10.1016/j.devcel.2024.11.002
Photosynthesis is a fundamental biological process for carbon fixation and capturing light energy to drive plant growth. However, excessive sunlight can cause photodamage, which is detrimental to plant health. To protect the photosynthetic machinery from over-excitation and subsequent damage, plant cells employ a sophisticated mechanism to regulate the distribution of excitation energy between the reaction centers and the antenna complexes—a process known as non-photochemical quenching (NPQ). The rapid relaxation of NPQ under low-light conditions and its swift induction in high-light environments are critical for preventing photodamage. However, the slow adjustment of NPQ to fluctuating light is estimated to reduce potential carbon gain by up to 40%. Accelerating the dynamic adjustment of NPQ to changing light conditions could, in principle, significantly enhance net photosynthetic efficiency. Using genome-wide association studies (GWAS), Feyissa and colleagues identified BOOSTER as a putative gene associated with NPQ in poplar. Overexpressing this gene increased photosynthetic efficiency and enhanced biomass production by up to 200% in both poplar and Arabidopsis. This discovery offers a promising strategy to improve plant productivity, with broad potential applications across various plant species. (Summary by Ching Chan @ntnuchanlab) Developmental Cell 10.1016/j.devcel.2024.11.002
A “GAME” changer in plant secondary metabolism
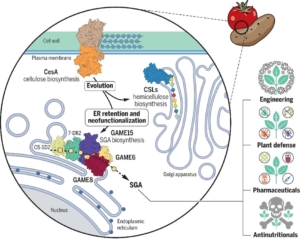 Cholesterol, an essential structural component of membranes and a precursor for steroid hormones, serves as a key metabolite at the interface of primary and secondary metabolism. However, the mechanisms regulating the balance between its diverse downstream metabolic pathways remain poorly understood. One well-known example of cholesterol-derived secondary metabolites is steroidal glycoalkaloids (SGAs). SGAs are potent plant defense compounds but can be self-toxic to plants above a certain threshold and act as antinutritional factors for humans. Understanding the biosynthesis of SGAs is, therefore, critical for balancing plant growth and defense while improving nutritional quality. In tomatoes, the GLYCOALKALOID METABOLISM (GAME) gene cluster has been linked to SGA biosynthesis. Two back-to-back studies, by Jozwiak et al. and Boccia et al., specifically investigated GAME15, which encodes an ER-localized cellulose synthase-like enzyme previously unassociated with SGA biosynthesis. Transient expression of GAME15 in Nicotiana benthamiana resulted in cholesterol glucuronidation and the accumulation of tomatidine, a precursor of SGA biosynthesis. Conversely, silencing GAME15 produced the opposite effect. Interestingly, phylogenetic studies revealed that the ancestral protein of GAME15, CesA, is plasma membrane-localized for cellulose fiber production. This is distinct from the role of GAME15 in cholesterol metabolism and SGA biosynthesis in the ER. These findings not only underscore the unique function of GAME15 as a regulatory hub for cholesterol metabolism but also highlight the evolutionary dynamics driving the emergence of novel metabolic pathways and metabolites. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.adq5721; and 10.1126/science.ado3409.
Cholesterol, an essential structural component of membranes and a precursor for steroid hormones, serves as a key metabolite at the interface of primary and secondary metabolism. However, the mechanisms regulating the balance between its diverse downstream metabolic pathways remain poorly understood. One well-known example of cholesterol-derived secondary metabolites is steroidal glycoalkaloids (SGAs). SGAs are potent plant defense compounds but can be self-toxic to plants above a certain threshold and act as antinutritional factors for humans. Understanding the biosynthesis of SGAs is, therefore, critical for balancing plant growth and defense while improving nutritional quality. In tomatoes, the GLYCOALKALOID METABOLISM (GAME) gene cluster has been linked to SGA biosynthesis. Two back-to-back studies, by Jozwiak et al. and Boccia et al., specifically investigated GAME15, which encodes an ER-localized cellulose synthase-like enzyme previously unassociated with SGA biosynthesis. Transient expression of GAME15 in Nicotiana benthamiana resulted in cholesterol glucuronidation and the accumulation of tomatidine, a precursor of SGA biosynthesis. Conversely, silencing GAME15 produced the opposite effect. Interestingly, phylogenetic studies revealed that the ancestral protein of GAME15, CesA, is plasma membrane-localized for cellulose fiber production. This is distinct from the role of GAME15 in cholesterol metabolism and SGA biosynthesis in the ER. These findings not only underscore the unique function of GAME15 as a regulatory hub for cholesterol metabolism but also highlight the evolutionary dynamics driving the emergence of novel metabolic pathways and metabolites. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.adq5721; and 10.1126/science.ado3409.
A fresh starch: Creating new starch granule morphologies in potato tuber
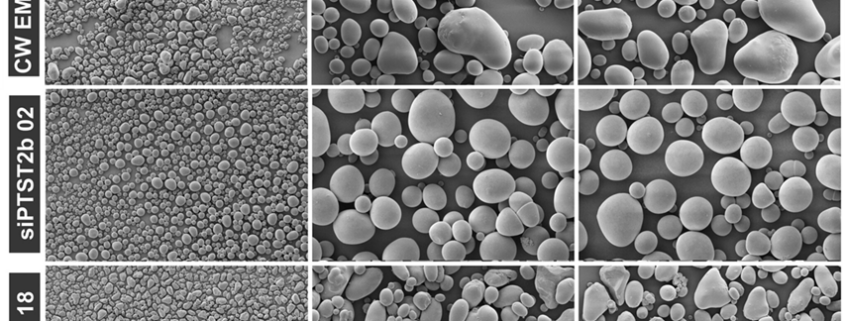
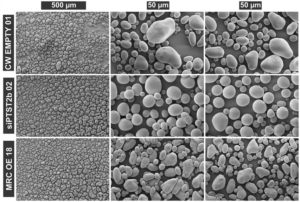 Starch is the major storage carbohydrate in plants and is organised into semicrystalline granules. The size, shape, and composition of these granules greatly affects how they are digested, and which industrial applications they are suitable for. Therefore, the enzymes controlling starch synthesis are incredibly valuable targets for breeders, despite being poorly understood outside of the model organism Arabidopsis. Hochmuth et al. investigated the role of two of these proteins in potato tubers: PROTEIN TARGETING TO STARCH2b (PTST2b) and MYOSIN RESEMBLING CHLOROPLAST PROTEIN (MRC). Tubers deficient in the newly-identified PTST2b paralogue due to silencing (referred to as siPTST2b plants) had granules that were the same size as the control, but were starkly spherical, rather than the wild-type ellipsoid shape. Notably, PTST2b did not interact with STARCH SYNTHASE 4, which is key to its function in Arabidopsis, suggesting these proteins may play different roles in tubers. MRC expression was high in the leaves but undetectable in tubers, and therefore its role was investigated using overexpression (MRC OE) lines. While potato starch granules typically are built from a single initiation point, the MRC OE lines had multiple initiation points per granule, and a smaller granule size than wild type tubers. Together, these potato mutants demonstrate the importance of PTST2b and MRC in starch granule morphology, and highlight the mechanistic differences in starch synthesis across different species and tissues. This work also generated two unique starch phenotypes which may be useful in the food industry. (Summary by Ciara O’Brien) Plant Biotechnol. J. 10.1111/pbi.14505
Starch is the major storage carbohydrate in plants and is organised into semicrystalline granules. The size, shape, and composition of these granules greatly affects how they are digested, and which industrial applications they are suitable for. Therefore, the enzymes controlling starch synthesis are incredibly valuable targets for breeders, despite being poorly understood outside of the model organism Arabidopsis. Hochmuth et al. investigated the role of two of these proteins in potato tubers: PROTEIN TARGETING TO STARCH2b (PTST2b) and MYOSIN RESEMBLING CHLOROPLAST PROTEIN (MRC). Tubers deficient in the newly-identified PTST2b paralogue due to silencing (referred to as siPTST2b plants) had granules that were the same size as the control, but were starkly spherical, rather than the wild-type ellipsoid shape. Notably, PTST2b did not interact with STARCH SYNTHASE 4, which is key to its function in Arabidopsis, suggesting these proteins may play different roles in tubers. MRC expression was high in the leaves but undetectable in tubers, and therefore its role was investigated using overexpression (MRC OE) lines. While potato starch granules typically are built from a single initiation point, the MRC OE lines had multiple initiation points per granule, and a smaller granule size than wild type tubers. Together, these potato mutants demonstrate the importance of PTST2b and MRC in starch granule morphology, and highlight the mechanistic differences in starch synthesis across different species and tissues. This work also generated two unique starch phenotypes which may be useful in the food industry. (Summary by Ciara O’Brien) Plant Biotechnol. J. 10.1111/pbi.14505
Unveiling REF1: A key regulator of plant regeneration
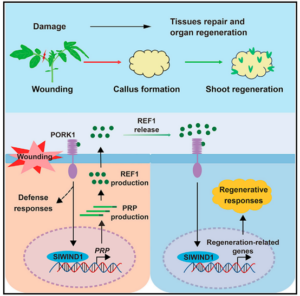 Plants frequently encounter damage during growth and development, necessitating remarkable regenerative abilities to repair damaged tissues. Plants can regenerate organs or even entire plants from callus or a single cell, a capacity underpinning asexual reproduction and various biotechnological applications. While numerous studies have investigated the plant regenerative response triggered by damage, the nature of the damage signal and its perception remain unclear. Tomato, a widely used model, has contributed significantly to the understanding of systemic defense and regeneration. Yang et al. characterized the tomato gene SPR9, which encodes the precursor of SlPep, which is a known elicitor peptide. The mutant spr9 exhibits impaired regenerative ability due to an attenuated damage signal critical for defense and regenerative responses. The study demonstrated that the loss of the SlPep precursor gene or its receptor disrupts wound-induced callus formation and the regenerative capacity, while its overexpression enhances regeneration. Furthermore, exogenous application of SlPep significantly improves regenerative capacity. Therefore, this small peptide has been renamed the regeneration factor REF1. REF1 activates SlWIND1, the main homolog of the tomato cell reprogramming regulator WIND1, which amplifies the REF1 signal by transcriptionally activating the REF1 precursor gene. In conclusion, this study established REF1 as a local wound signal for plant regeneration. This discovery provides a promising target for improving the regenerative capacity and transformation efficiency of recalcitrant crops. (Summary by Yuanyuan Liu @YuanyuanLiu12) Cell 10.1016/j.cell.2024.04.040
Plants frequently encounter damage during growth and development, necessitating remarkable regenerative abilities to repair damaged tissues. Plants can regenerate organs or even entire plants from callus or a single cell, a capacity underpinning asexual reproduction and various biotechnological applications. While numerous studies have investigated the plant regenerative response triggered by damage, the nature of the damage signal and its perception remain unclear. Tomato, a widely used model, has contributed significantly to the understanding of systemic defense and regeneration. Yang et al. characterized the tomato gene SPR9, which encodes the precursor of SlPep, which is a known elicitor peptide. The mutant spr9 exhibits impaired regenerative ability due to an attenuated damage signal critical for defense and regenerative responses. The study demonstrated that the loss of the SlPep precursor gene or its receptor disrupts wound-induced callus formation and the regenerative capacity, while its overexpression enhances regeneration. Furthermore, exogenous application of SlPep significantly improves regenerative capacity. Therefore, this small peptide has been renamed the regeneration factor REF1. REF1 activates SlWIND1, the main homolog of the tomato cell reprogramming regulator WIND1, which amplifies the REF1 signal by transcriptionally activating the REF1 precursor gene. In conclusion, this study established REF1 as a local wound signal for plant regeneration. This discovery provides a promising target for improving the regenerative capacity and transformation efficiency of recalcitrant crops. (Summary by Yuanyuan Liu @YuanyuanLiu12) Cell 10.1016/j.cell.2024.04.040
Spatially resolved, single-cell multi-omics atlas of soybean development
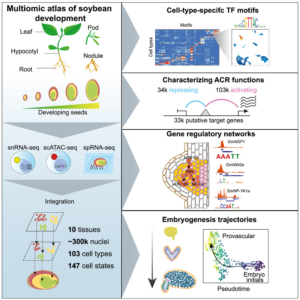 In this exciting paper, Zhang, Luo, Marand et al. combined several powerful techniques to investigate the program underlying soybean seed development. They used RNA sequencing to profile gene expression from single nuclei (snRNA-seq). They also carried out single-cell sequencing of assays for transposase-accessible chromatin (scATAC-seq) to identify accessible chromatin regions (ACR), which are potentially regions where transcription factors can bind. Combining these datasets with laser-capture microdissection RNA-seq datasets allowed the authors to pinpoint atlas-scale cell-type regulatory regions and gene expression profiles across 10 developmental stages and tissues. In addition to sharing these datasets in the Soybean Multi-Omics Atlas (https://soybean-atlas.com/), the authors share the first of surely many new insights. For example, they identified subtypes of endosperm cells with distinct patterns of expression of nutrient transporters and cell death, indicating a temporal as well as spatial pattern of nutrient support of the developing embryo. The analysis of the accessible chromatin regions (ACRs) is particularly interesting, as the authors found a high cell-type specific correlation between gene accessiblity and gene expression, and identified key transcription factors that define cell types; perhaps not surprising, but nevertheless intriguing. As the authors state, “We anticipate that the real potential of single-cell methods will extend beyond aiding gene function studies and uncovering regulatory networks.” Clearly these data provide a wealth of information to explore and analyze. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Cell 10.1016/j.cell.2024.10.050
In this exciting paper, Zhang, Luo, Marand et al. combined several powerful techniques to investigate the program underlying soybean seed development. They used RNA sequencing to profile gene expression from single nuclei (snRNA-seq). They also carried out single-cell sequencing of assays for transposase-accessible chromatin (scATAC-seq) to identify accessible chromatin regions (ACR), which are potentially regions where transcription factors can bind. Combining these datasets with laser-capture microdissection RNA-seq datasets allowed the authors to pinpoint atlas-scale cell-type regulatory regions and gene expression profiles across 10 developmental stages and tissues. In addition to sharing these datasets in the Soybean Multi-Omics Atlas (https://soybean-atlas.com/), the authors share the first of surely many new insights. For example, they identified subtypes of endosperm cells with distinct patterns of expression of nutrient transporters and cell death, indicating a temporal as well as spatial pattern of nutrient support of the developing embryo. The analysis of the accessible chromatin regions (ACRs) is particularly interesting, as the authors found a high cell-type specific correlation between gene accessiblity and gene expression, and identified key transcription factors that define cell types; perhaps not surprising, but nevertheless intriguing. As the authors state, “We anticipate that the real potential of single-cell methods will extend beyond aiding gene function studies and uncovering regulatory networks.” Clearly these data provide a wealth of information to explore and analyze. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Cell 10.1016/j.cell.2024.10.050
Key regulators of juvenile-to-adult phase change
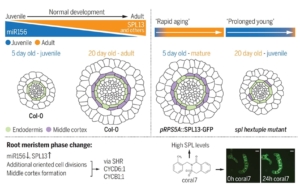 The precise control of cell division orientation drives plant 3D structure formation, enabling radial and longitudinal growth. The SPL pathway is closely linked to age-related processes in the shoot, driving the vegetative transition from juvenile to adult phases by regulating specific morphological and molecular traits. Similarly, in the root apical meristem (RAM), SPL transcription factors orchestrate a comparable phase transition, highlighting their pivotal role in developmental shifts across both shoot and root systems. To identify key regulators, Yang et al. screened over 15,000 compounds using a high-throughput confocal imaging system. They identified a compound, coral7, that induces changes in cell division orientation by promoting expression of the transcription factor SPL13. The research team analyzed RAM development over time and found that late-stage roots undergo significant morphological changes, including middle cortex formation and meristem thickening, driven by reoriented cell divisions. SPL transcription factors were shown to regulate these processes by activating key cell division-related genes, such as CYCB1 and CYCD6;1, through the SHR pathway. Additionally, studies on monocot rice and its related spl mutants similarly revealed that SPL plays a key role in cell division orientation and root thickening in the rice root meristem. This demonstrates that SPL is a conserved gene responsible for remodeling root morphology (thickness) across plants. (Summary by Yuanyuan Liu @YuanyuanLiu12) Science 10.1126/science.ado4298
The precise control of cell division orientation drives plant 3D structure formation, enabling radial and longitudinal growth. The SPL pathway is closely linked to age-related processes in the shoot, driving the vegetative transition from juvenile to adult phases by regulating specific morphological and molecular traits. Similarly, in the root apical meristem (RAM), SPL transcription factors orchestrate a comparable phase transition, highlighting their pivotal role in developmental shifts across both shoot and root systems. To identify key regulators, Yang et al. screened over 15,000 compounds using a high-throughput confocal imaging system. They identified a compound, coral7, that induces changes in cell division orientation by promoting expression of the transcription factor SPL13. The research team analyzed RAM development over time and found that late-stage roots undergo significant morphological changes, including middle cortex formation and meristem thickening, driven by reoriented cell divisions. SPL transcription factors were shown to regulate these processes by activating key cell division-related genes, such as CYCB1 and CYCD6;1, through the SHR pathway. Additionally, studies on monocot rice and its related spl mutants similarly revealed that SPL plays a key role in cell division orientation and root thickening in the rice root meristem. This demonstrates that SPL is a conserved gene responsible for remodeling root morphology (thickness) across plants. (Summary by Yuanyuan Liu @YuanyuanLiu12) Science 10.1126/science.ado4298
Abundant, unusual RNAs on the leaf surface
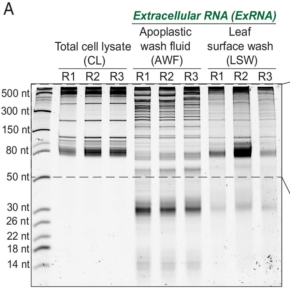 It seems that there is no end to the surprises that RNA provides. To the old-school trio of mRNA, tRNA, and rRNA, the past 20 years have added miRNA, siRNA, tasiRNA and others, all with unique and specific roles in regulating gene activity. More recently, evidence has been accumulating that demonstrates non-cell autonomous RNA activities including as facilitators of plant defense. For example, in host-induced gene silencing, plants express a transgene encoding a double-stranded RNA that silences specific pathogen genes. Recent identification of abundant endogenously-produced extracellular RNAs (exRNAs) has led to several questions and hypotheses about their origin and function. Previous studies have looked at apoplastic RNAs (located between cells for example in cell walls) and RNAs encapsulated in extracellular vesicles (EVs). In this new work, Borniego and Singla-Rastogi et al. have characterized another pool of exRNAs, those abundant on the leaf surface but not encapsulated in vesicles. The authors carefully analyzed these RNAs and found that they are a unique population, distinct from those within the cell, within the apoplast, or within the EVs. The leaf surface RNAs are largely plant derived, mainly from tRNAs but other RNA classes are also represented. The authors speculate on their origin (maybe released from trichomes?) and suggest that they have a role in shaping the leaf microbiome. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2409090121
It seems that there is no end to the surprises that RNA provides. To the old-school trio of mRNA, tRNA, and rRNA, the past 20 years have added miRNA, siRNA, tasiRNA and others, all with unique and specific roles in regulating gene activity. More recently, evidence has been accumulating that demonstrates non-cell autonomous RNA activities including as facilitators of plant defense. For example, in host-induced gene silencing, plants express a transgene encoding a double-stranded RNA that silences specific pathogen genes. Recent identification of abundant endogenously-produced extracellular RNAs (exRNAs) has led to several questions and hypotheses about their origin and function. Previous studies have looked at apoplastic RNAs (located between cells for example in cell walls) and RNAs encapsulated in extracellular vesicles (EVs). In this new work, Borniego and Singla-Rastogi et al. have characterized another pool of exRNAs, those abundant on the leaf surface but not encapsulated in vesicles. The authors carefully analyzed these RNAs and found that they are a unique population, distinct from those within the cell, within the apoplast, or within the EVs. The leaf surface RNAs are largely plant derived, mainly from tRNAs but other RNA classes are also represented. The authors speculate on their origin (maybe released from trichomes?) and suggest that they have a role in shaping the leaf microbiome. (Summary by Mary Williams @PlantTeaching.bksy.social @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2409090121