Plant Science Research Weekly: February 8th
Opinion: To learn inclusion skills, make it personal
 This is a great essay by David Asai, Senior Director for science education at the Howard Hughes Medical Institute. David describes his journey from feeling annoyed at having to attend a multicultural forum to embracing their merit. He describes exercises that promote empathy and encourage respectful listening. He writes, “We understand that scientific excellence depends on creativity, that creativity emerges from diversity, and that the advantages of diversity are realized through inclusion,” and “Inclusion is a feeling of belonging, and so creating an empowering, embracing, egalitarian environment starts with the heart.” Over the years I have learned a lot from working with David, and I am pleased to note that he will be speaking at the Plant Biology 2019 conference, in the session “Biological and Personal Networks: Why They Matter for Plant Biology”. (Summary by Mary Williams) Nature 10.1038/d41586-019-00282-y
This is a great essay by David Asai, Senior Director for science education at the Howard Hughes Medical Institute. David describes his journey from feeling annoyed at having to attend a multicultural forum to embracing their merit. He describes exercises that promote empathy and encourage respectful listening. He writes, “We understand that scientific excellence depends on creativity, that creativity emerges from diversity, and that the advantages of diversity are realized through inclusion,” and “Inclusion is a feeling of belonging, and so creating an empowering, embracing, egalitarian environment starts with the heart.” Over the years I have learned a lot from working with David, and I am pleased to note that he will be speaking at the Plant Biology 2019 conference, in the session “Biological and Personal Networks: Why They Matter for Plant Biology”. (Summary by Mary Williams) Nature 10.1038/d41586-019-00282-y
News and Views: Harnessing the potential of germplasm collections
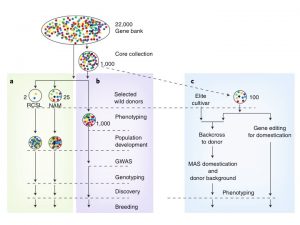 There are more than 7 million crop accessions in gene banks around the world, however the information records for them are in multiple formats, making a comparison between and within collections difficult. Extrapolating from a new paper by Milner et al. (Nature Genetics 51: pages319–326) Langridge and Waugh suggest a framework to efficiently utilize this genetic diversity in crop improvement. The first step will be to characterize the collection by low cost high throughput genotyping technology which will allow the identification of duplicates, geographical structures and mix-ups. With this common information to all accessions, a representative core can be selected for deeper characterization. The second approach recommended is the use of contemporary breeding methods like CRISPR-Cas to introduce important agriculture genes already identified in elite lines into diverse germplasm. This strategy will help circumvent the problem of low recombination rates between wild relatives and elite lines in crops with a large genome. (Summary by Celilia Vasquez-Robinet) Nat. Genet. 10.1038/s41588-018-0340-4
There are more than 7 million crop accessions in gene banks around the world, however the information records for them are in multiple formats, making a comparison between and within collections difficult. Extrapolating from a new paper by Milner et al. (Nature Genetics 51: pages319–326) Langridge and Waugh suggest a framework to efficiently utilize this genetic diversity in crop improvement. The first step will be to characterize the collection by low cost high throughput genotyping technology which will allow the identification of duplicates, geographical structures and mix-ups. With this common information to all accessions, a representative core can be selected for deeper characterization. The second approach recommended is the use of contemporary breeding methods like CRISPR-Cas to introduce important agriculture genes already identified in elite lines into diverse germplasm. This strategy will help circumvent the problem of low recombination rates between wild relatives and elite lines in crops with a large genome. (Summary by Celilia Vasquez-Robinet) Nat. Genet. 10.1038/s41588-018-0340-4
The global burden of pathogens and pests on major food crops
 Pathogens and pests are bad, but just how bad? And how do different regions of the world compare in terms of crop losses to pathogens and pests? Savary et al. surveyed crop experts from across the globe to address these questions, focusing on five major food crops (wheat, rice, maize, potato and soybean). They identify where some of the most detrimental crop/ pest interactions take place. They conclude that “the highest losses are associated with food-deficit regions with fast-growing populations, and frequently with emerging or re-emerging pests and diseases.” The data are interesting to browse and certainly will be useful for identifying focus areas for research and intervention. (Summary by Mary Williams) Nature Eco. Evol. 10.1038/s41559-018-0793-y
Pathogens and pests are bad, but just how bad? And how do different regions of the world compare in terms of crop losses to pathogens and pests? Savary et al. surveyed crop experts from across the globe to address these questions, focusing on five major food crops (wheat, rice, maize, potato and soybean). They identify where some of the most detrimental crop/ pest interactions take place. They conclude that “the highest losses are associated with food-deficit regions with fast-growing populations, and frequently with emerging or re-emerging pests and diseases.” The data are interesting to browse and certainly will be useful for identifying focus areas for research and intervention. (Summary by Mary Williams) Nature Eco. Evol. 10.1038/s41559-018-0793-y
I see the light! Fluorescent proteins suitable for cell wall/apoplast targeting in Nicotiana benthamiana leaves
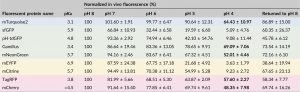 Imaging proteins through fluorescent tagging is tremendously powerful but also tricky. In this very useful paper, Stottard and Rolland survey the properties of 10 fluorescent tags to determine their effectiveness in the low pH environment of the plant cell wall and apoplast. The authors also test the propensity for the fluorescent proteins to aggregate, and whether or not they can be targeted to the apoplast. Finally, they describe parameters to consider when choosing a protein tag. A great read before you plan your next study! (Summary by Mary Williams) Plant Direct 10.1002/pld3.112
Imaging proteins through fluorescent tagging is tremendously powerful but also tricky. In this very useful paper, Stottard and Rolland survey the properties of 10 fluorescent tags to determine their effectiveness in the low pH environment of the plant cell wall and apoplast. The authors also test the propensity for the fluorescent proteins to aggregate, and whether or not they can be targeted to the apoplast. Finally, they describe parameters to consider when choosing a protein tag. A great read before you plan your next study! (Summary by Mary Williams) Plant Direct 10.1002/pld3.112
Proteome-wide, structure-based prediction of protein-protein interactions
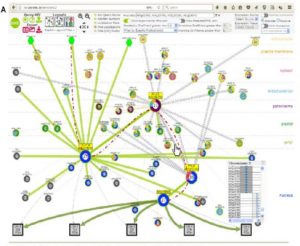 Genomics and transcriptomics have brought huge advances in understanding of plant science, but proteomics is both more challenging and in some ways more relevant to understand what is happening inside of a cell. Proteins function largely through their interactions with other proteins, so it is important to be able to confidently predict protein-protein interactions. Dong et al. describe a molecular interactions viewer that builds upon demonstrated structural and interaction data to predict protein-protein interactions. The authors predict new interactions which they verify using yeast-two hybrid studies. Their predicted structure-ome is freely available through the Bio-Analytic Resource (BAR) as the Arabidopsis Interactions Viewer 2 (http://bar.utoronto.ca/interactions2/). (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01216.
Genomics and transcriptomics have brought huge advances in understanding of plant science, but proteomics is both more challenging and in some ways more relevant to understand what is happening inside of a cell. Proteins function largely through their interactions with other proteins, so it is important to be able to confidently predict protein-protein interactions. Dong et al. describe a molecular interactions viewer that builds upon demonstrated structural and interaction data to predict protein-protein interactions. The authors predict new interactions which they verify using yeast-two hybrid studies. Their predicted structure-ome is freely available through the Bio-Analytic Resource (BAR) as the Arabidopsis Interactions Viewer 2 (http://bar.utoronto.ca/interactions2/). (Summary by Mary Williams) Plant Physiol. 10.1104/pp.18.01216.
Rubisco condensate formation by CcmM in β-carboxysome biogenesis ($)
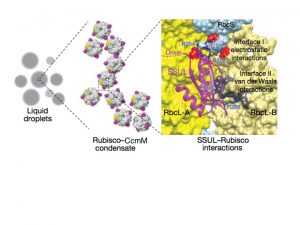 Cyanobacterial carbon-dioxide concentrating mechanisms elevate intracellular inorganic carbon as bicarbonate, and then concentrate it as carbon-dioxide around the enzyme Rubisco in specialized protein micro-compartments called carboxysomes. The formation of B-carboxysomes involves an aggregation between Rubisco and the major scaffolding protein of the carboxysome, called CcmM. CcmM is composed of several repeat modules that can resemble the small subunit of Rubisco, and it was long assumed that they replaced these small subunits of Rubisco when the proteins interact, linking Rubisco proteins within the carboxysome. However, the highly dynamic complex formed between CcmM and Rubisco has made it difficult to determine the structural formation by traditional structural biology methods. Wang et al. used purified Rubisco and CcmM from Synechococcus elongates to explore the structural relationship between these two proteins, and unexpectedly found that they form a spontaneous aggregate, via liquid-like phase separation. Using the turbidity of this condensate and variants of the CcmM and Rubisco proteins, they were able to determine the regions of importance, and then resolved the structure basis of the interaction using cryo-electron microscopy to find that the small-subunit like modules of the CcmM bind close to the equatorial region of Rubisco between large subunit dimers, but also interact with the Rubisco small subunit, ensuring that only the intact holoenzyme is incorporated into the B-carboxysome. (Summary by Amanda Cavanagh) Nature 10.1038/s41586-019-0880-5
Cyanobacterial carbon-dioxide concentrating mechanisms elevate intracellular inorganic carbon as bicarbonate, and then concentrate it as carbon-dioxide around the enzyme Rubisco in specialized protein micro-compartments called carboxysomes. The formation of B-carboxysomes involves an aggregation between Rubisco and the major scaffolding protein of the carboxysome, called CcmM. CcmM is composed of several repeat modules that can resemble the small subunit of Rubisco, and it was long assumed that they replaced these small subunits of Rubisco when the proteins interact, linking Rubisco proteins within the carboxysome. However, the highly dynamic complex formed between CcmM and Rubisco has made it difficult to determine the structural formation by traditional structural biology methods. Wang et al. used purified Rubisco and CcmM from Synechococcus elongates to explore the structural relationship between these two proteins, and unexpectedly found that they form a spontaneous aggregate, via liquid-like phase separation. Using the turbidity of this condensate and variants of the CcmM and Rubisco proteins, they were able to determine the regions of importance, and then resolved the structure basis of the interaction using cryo-electron microscopy to find that the small-subunit like modules of the CcmM bind close to the equatorial region of Rubisco between large subunit dimers, but also interact with the Rubisco small subunit, ensuring that only the intact holoenzyme is incorporated into the B-carboxysome. (Summary by Amanda Cavanagh) Nature 10.1038/s41586-019-0880-5
What quantity of photosystem I is optimum for safe photosynthesis? ($)
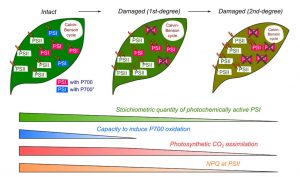 Photosynthesis is one of the most vital and complex processes carried out by plants. During photosynthesis, Photosystem I and II (PSI and PSII) undergo photo-excitation. Excessive photo-excitation can damage and deactivate the PSI by generating reactive oxygen species (ROS). This kind of photoinhibition of PSI is a lethal event that can abruptly inhibit the photosynthesis and growth of oxygenic photoautotrophs. However, photoinhibition of PSI in plants does not occur regularly as plants control it by maintaining the PSI at its oxidative state. Observations from different studies suggest that the optimized quantity of PSI can induce protection against photoinhibition and ensure robust photosynthesis. Shimakawa and Miyake characterized the effects of photochemically active and inactivated PSI on a variety of photosynthetic parameters such as oxidation of P700, photosynthetic CO2 assimilation, and interrelation of PSI and PSII, using intact leaves of the Arabidopsis thaliana. The active and inactivated state of PSI was maintained by using repetitive short pulse (rSP) illumination. They observed that there is a linear correlation between PSI activity and P700 oxidation and CO2 assimilation for a range of 100% to 60% of PSI activity. (Summary by Mugdha Sabale) Plant Physiol. 10.1104/pp.18.01493
Photosynthesis is one of the most vital and complex processes carried out by plants. During photosynthesis, Photosystem I and II (PSI and PSII) undergo photo-excitation. Excessive photo-excitation can damage and deactivate the PSI by generating reactive oxygen species (ROS). This kind of photoinhibition of PSI is a lethal event that can abruptly inhibit the photosynthesis and growth of oxygenic photoautotrophs. However, photoinhibition of PSI in plants does not occur regularly as plants control it by maintaining the PSI at its oxidative state. Observations from different studies suggest that the optimized quantity of PSI can induce protection against photoinhibition and ensure robust photosynthesis. Shimakawa and Miyake characterized the effects of photochemically active and inactivated PSI on a variety of photosynthetic parameters such as oxidation of P700, photosynthetic CO2 assimilation, and interrelation of PSI and PSII, using intact leaves of the Arabidopsis thaliana. The active and inactivated state of PSI was maintained by using repetitive short pulse (rSP) illumination. They observed that there is a linear correlation between PSI activity and P700 oxidation and CO2 assimilation for a range of 100% to 60% of PSI activity. (Summary by Mugdha Sabale) Plant Physiol. 10.1104/pp.18.01493
Leaf age dictates abiotic versus biotic stress signalling in Arabidopsis
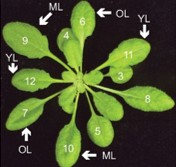 Plants sense and respond to various external stimuli throughout their lifespan. During stress responses brought forth by abiotic or biotic factors, molecular and physiological adjustments mediated by distinct yet interconnected hormone pathways play critical roles in plant survival. Berens et al. investigate how leaves of the Arabidopsis rosette prioritize stress signalling outputs during combined or sequential abiotic and biotic stress treatments. The authors uncover a differential and antagonistic prioritization of abscisic acid (ABA)-mediated abiotic stress signalling versus salicylic acid (SA)-mediated biotic signalling in older leaves of the rosette. Transcriptomic and directed expression studies further expanded on this paradigm, revealing strong ABA-mediated suppression of SA-dependent immune signalling in older Arabidopsis leaves that ultimately results in increased susceptibility to the hemibiotrophic bacterial pathogen Pseudomonas syringae. By contrast, younger Arabidopsis leaves are insensitive to ABA-induced immune suppression and thus remain more resistant to infection in a manner dependent on the SA signalling and accumulation-regulating enzyme PBS3. Further phenotypic analyses demonstrate that Arabidopsis PBS3 plays critical roles in maintaining reproductive success under combined stress and in shaping the age and stress-dependent microbial communities of rosette leaves. Collectively, this work reveals new layers of complexity in the regulation of stress signalling networks that have evolved to differentially protect leaves throughout their development. (Summary by Phil Carella) Proc. Natl. Acad. Sci. USA
Plants sense and respond to various external stimuli throughout their lifespan. During stress responses brought forth by abiotic or biotic factors, molecular and physiological adjustments mediated by distinct yet interconnected hormone pathways play critical roles in plant survival. Berens et al. investigate how leaves of the Arabidopsis rosette prioritize stress signalling outputs during combined or sequential abiotic and biotic stress treatments. The authors uncover a differential and antagonistic prioritization of abscisic acid (ABA)-mediated abiotic stress signalling versus salicylic acid (SA)-mediated biotic signalling in older leaves of the rosette. Transcriptomic and directed expression studies further expanded on this paradigm, revealing strong ABA-mediated suppression of SA-dependent immune signalling in older Arabidopsis leaves that ultimately results in increased susceptibility to the hemibiotrophic bacterial pathogen Pseudomonas syringae. By contrast, younger Arabidopsis leaves are insensitive to ABA-induced immune suppression and thus remain more resistant to infection in a manner dependent on the SA signalling and accumulation-regulating enzyme PBS3. Further phenotypic analyses demonstrate that Arabidopsis PBS3 plays critical roles in maintaining reproductive success under combined stress and in shaping the age and stress-dependent microbial communities of rosette leaves. Collectively, this work reveals new layers of complexity in the regulation of stress signalling networks that have evolved to differentially protect leaves throughout their development. (Summary by Phil Carella) Proc. Natl. Acad. Sci. USA
Global topological order emerges through local mechanical control of cell divisions in the Arabidopsis shoot apical meristem

In plants, the final shape of organs depends on how and when the cells divide. To get some insights about the rules governing this process, Jackson et al. applied network science to study the cell organization dynamics in the shoot apical meristem (SAM). They studied the L1, L2 and L3 cell layers in the SAM, where they tracked cell division over time. Cells in that tissue can be represented as a spatial network, in this case, nodes represent cells, and edges are the shared cell interfaces. Once the networks are generated, topological analysis can be performed as in other kinds of networks. The authors analyzed how local (i.e., node degree) and global (i.e., betweenness centrality) topological properties are related to cell division. The implications of mechanical interactions between cells were also studied using the katanin1 mutant, in which microtubule networks are disrupted, affecting mechanical interactions between cells. This work is helping to understand how cells organize in order to create complex tissues and organs. (Summary by Humberto Herrera-Umbaldo) Cell Systems 10.1016/j.cels.2018.12.009
Endogenous control of root system architecture through hypoxic conditions ($)
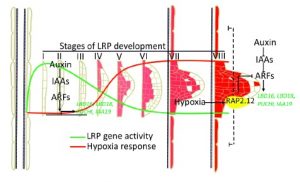 The three-dimensional structure of plant roots, often referred to as the root system architecture (RSA), is important for soil exploration and nutrient acquisition. The regulation of the RSA is important to prevent overuse of resources and to ensure efficient function. A recent study from Shukla et al. found that regulation of RSA occurs through the formation of endogenous hypoxic (low oxygen) niches and the inhibition of new lateral roots in Arabidopsis thaliana. Using available transcription data and mutant lines, the authors found that localized hypoxic niches are formed during lateral root development and induce a family of transcription factors commonly associated with hypoxic response, ERF-VII. Furthermore, the ERF-VII family repress the activation of lateral root associated genes, preventing their positive regulation in the presence of auxin. Taken together, the results indicate that RSA regulation is controlled by the plant itself by establishing hypoxic niches and repressing genes that are critical for lateral root formation. (Summary by Nick Segerson) Mol. Plant 10.1016/j.molp.2019.01.007
The three-dimensional structure of plant roots, often referred to as the root system architecture (RSA), is important for soil exploration and nutrient acquisition. The regulation of the RSA is important to prevent overuse of resources and to ensure efficient function. A recent study from Shukla et al. found that regulation of RSA occurs through the formation of endogenous hypoxic (low oxygen) niches and the inhibition of new lateral roots in Arabidopsis thaliana. Using available transcription data and mutant lines, the authors found that localized hypoxic niches are formed during lateral root development and induce a family of transcription factors commonly associated with hypoxic response, ERF-VII. Furthermore, the ERF-VII family repress the activation of lateral root associated genes, preventing their positive regulation in the presence of auxin. Taken together, the results indicate that RSA regulation is controlled by the plant itself by establishing hypoxic niches and repressing genes that are critical for lateral root formation. (Summary by Nick Segerson) Mol. Plant 10.1016/j.molp.2019.01.007
Oscillating aquaporin phosphorylations and 14-3-3 proteins mediate circadian regulation of leaf hydraulics
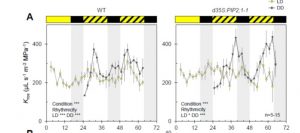 Plant activities change drastically between the light and dark times of day. Many of these different activities can be observed through transcriptional studies, which show that gene expression goes up and down over the course of the day. Many of these transcriptional changes are driven by the circadian clock. Here Prado et al. show that transport of water in leaves (hydraulic conductivity) also follows circadian patterns, and that these patterns are in part driven by post-translational modifications of the aquaporin channel protein AtPIP2;1. These clock-driven cycles of phosphorylation / dephosphorylation may affect interactions with 14-3-3 proteins. Understanding clock-regulated leaf water status may lead to improvements in crop performance. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00804
Plant activities change drastically between the light and dark times of day. Many of these different activities can be observed through transcriptional studies, which show that gene expression goes up and down over the course of the day. Many of these transcriptional changes are driven by the circadian clock. Here Prado et al. show that transport of water in leaves (hydraulic conductivity) also follows circadian patterns, and that these patterns are in part driven by post-translational modifications of the aquaporin channel protein AtPIP2;1. These clock-driven cycles of phosphorylation / dephosphorylation may affect interactions with 14-3-3 proteins. Understanding clock-regulated leaf water status may lead to improvements in crop performance. (Summary by Mary Williams) Plant Cell 10.1105/tpc.18.00804
A silk-expressed pectin methylesterase confers cross-incompatibility between wild and domesticated strains of Zea mays
 The key event for speciation is reproductive isolation. In sexually reproducing plants, genetic factors act as reproductive barriers preventing the interbreeding of related strains. In this study, Lu et al. aimed to explore how the Tcb1-s (Teosinte crossing barrier 1-s) haplotype, found exclusively in wild teosinte, blocks maize pollen fertilization. Using mixed pollination tests with knockout mutants, as well as genetic and genomic data, the authors demonstrate the Tcb1-s has functional male and female genes that enable self-type fertilization, but prevent pollination by pollen carrying the tcb1 haplotype found in almost all maize lines. The results reveal the barrier role of the female gene in Tcb1-s lines, since it encodes a pectin methylesterase expressed by the pistil, which affects the cell wall of the pollen tube and compromises fertilization. This work creates a paradigm for future studies of speciation mechanisms in other plants, which may be useful to manage pollen contamination for crop improvement techniques. (Summary by Ana Valladares) bioRxiv
The key event for speciation is reproductive isolation. In sexually reproducing plants, genetic factors act as reproductive barriers preventing the interbreeding of related strains. In this study, Lu et al. aimed to explore how the Tcb1-s (Teosinte crossing barrier 1-s) haplotype, found exclusively in wild teosinte, blocks maize pollen fertilization. Using mixed pollination tests with knockout mutants, as well as genetic and genomic data, the authors demonstrate the Tcb1-s has functional male and female genes that enable self-type fertilization, but prevent pollination by pollen carrying the tcb1 haplotype found in almost all maize lines. The results reveal the barrier role of the female gene in Tcb1-s lines, since it encodes a pectin methylesterase expressed by the pistil, which affects the cell wall of the pollen tube and compromises fertilization. This work creates a paradigm for future studies of speciation mechanisms in other plants, which may be useful to manage pollen contamination for crop improvement techniques. (Summary by Ana Valladares) bioRxiv
Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin
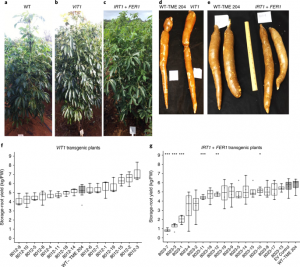 There are many forms of hunger, one of which is micronutrient deficiency. Cassava is a staple food, but low in iron and zinc, in many regions where deficiencies of these micronutrients are common; therefore efforts have been made to biofortify cassava. Traditional breeding methods are of limited value as there is little genetic diversity in cassava for iron and zinc accumulation, so transgenic approaches may be more effective. By introducing genes encoding an iron transporter (IRT1) and ferritin (FER1), Narayanan et al. were able to produce cassava that “accumulated iron levels 7–18 times higher and zinc levels 3–10 times higher than those in nontransgenic controls in the field” without negatively affecting shoot and root biomass. (Summary by Mary Williams) Nature Biotech. 10.1038/s41587-018-0002-1
There are many forms of hunger, one of which is micronutrient deficiency. Cassava is a staple food, but low in iron and zinc, in many regions where deficiencies of these micronutrients are common; therefore efforts have been made to biofortify cassava. Traditional breeding methods are of limited value as there is little genetic diversity in cassava for iron and zinc accumulation, so transgenic approaches may be more effective. By introducing genes encoding an iron transporter (IRT1) and ferritin (FER1), Narayanan et al. were able to produce cassava that “accumulated iron levels 7–18 times higher and zinc levels 3–10 times higher than those in nontransgenic controls in the field” without negatively affecting shoot and root biomass. (Summary by Mary Williams) Nature Biotech. 10.1038/s41587-018-0002-1
Introducing curcumin biosynthesis in Arabidopsis enhances lignocellulosic biomass processing
 Lignin, polymerized from aromatic monolignols, provides strength to cell walls but its resistance to enzymatic degradation thwarts efforts to isolate cellulosic carbohydrates from cell walls for biofuels and other applications. It has been shown that lignin’s properties can be altered by the incorporation of other monomers. Here, Oyarce et al. have introduced genes for the synthesis of curcumin (from turmeric, Curcuma longa), which is structurally similar to the monolignol coniferyl ferulate, into Arabidopsis, and shown that the curcumin is incorporated into the lignin polymer. Curcumin introduces an alkali-labile bond into the lignin, providing a way to more easily remove lignin from plant tissues. The authors show that stem biomass and strength are not affected by the incorporation of curcumin, but that after akalki treatment, glucose release from the engineered plants is significantly increased. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0350-3
Lignin, polymerized from aromatic monolignols, provides strength to cell walls but its resistance to enzymatic degradation thwarts efforts to isolate cellulosic carbohydrates from cell walls for biofuels and other applications. It has been shown that lignin’s properties can be altered by the incorporation of other monomers. Here, Oyarce et al. have introduced genes for the synthesis of curcumin (from turmeric, Curcuma longa), which is structurally similar to the monolignol coniferyl ferulate, into Arabidopsis, and shown that the curcumin is incorporated into the lignin polymer. Curcumin introduces an alkali-labile bond into the lignin, providing a way to more easily remove lignin from plant tissues. The authors show that stem biomass and strength are not affected by the incorporation of curcumin, but that after akalki treatment, glucose release from the engineered plants is significantly increased. (Summary by Mary Williams) Nature Plants 10.1038/s41477-018-0350-3
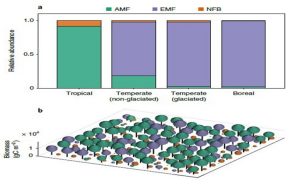 Most plants depend on microbial partners (symbionts) to help them take up nutrients from the soil. Lu and Hedin set out to identify how these plant-symbiont partnerships contribute to plant distributions and biogeochemical cycles, using an evolution-based modelling approach. The model takes into account forest carbon and nutrient cycles, nutrient limitations and the exchange of photosynthetically acquired carbon for nutrients gained from belowground symbionts. The manner in which this method differs is that enables the most successful individual plant strategies to emerge within the plant-nutrient cycle, regardless of whether the exposed properties are optimal for the ecosystem. Based on results, a modelled world without symbioses would have a 48 and 19 % drop in tropical and boreal forests when it comes to nitrogen and phosphorus cycling. That leads to the conclusion that the evolution of belowground symbioses influenced the cycles discussed in the paper. (Summary by Tina Ugulin) Nature Ecol. Evol. 10.1038/s41559-018-0759-0
Most plants depend on microbial partners (symbionts) to help them take up nutrients from the soil. Lu and Hedin set out to identify how these plant-symbiont partnerships contribute to plant distributions and biogeochemical cycles, using an evolution-based modelling approach. The model takes into account forest carbon and nutrient cycles, nutrient limitations and the exchange of photosynthetically acquired carbon for nutrients gained from belowground symbionts. The manner in which this method differs is that enables the most successful individual plant strategies to emerge within the plant-nutrient cycle, regardless of whether the exposed properties are optimal for the ecosystem. Based on results, a modelled world without symbioses would have a 48 and 19 % drop in tropical and boreal forests when it comes to nitrogen and phosphorus cycling. That leads to the conclusion that the evolution of belowground symbioses influenced the cycles discussed in the paper. (Summary by Tina Ugulin) Nature Ecol. Evol. 10.1038/s41559-018-0759-0