Plant Science Research Weekly: February 7, 2025
Review: Genetic engineering for carbon assimilation in plants
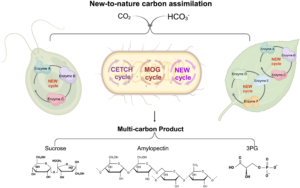 Rubisco (Ribulose‐1,5‐bisphosphate carboxylase/oxygenase) is the central enzyme for photosynthesis, This enzyme poorly discriminates between CO2 and O2, which limits its efficiency. To work around this and make carbon assimilation more efficient, scientists have been employing different engineering strategies, several of which are summarized in a new review by Qin et al. One strategy is to alter Rubisco activity, for example by inducing mutations, combining parts of Rubisco from different organisms, engineering other enzymes involved with Rubisco activation and accumulation, and introducing Rubisco genes from lower organisms to crop plants. Another engineering approach is to create carbon-concentrating compartments, for example by using carboxysomes found in cyanobacteria and pyrenoids from eukaryotic algae. Alternatively, CO2 levels at Rubisco can be boosted by introducing the C4 cycle in C3 plants. Scientists are also engineering the Calvin-Benson-Bassham Cycle by overexpressing specific enzymes involved in the cycle, as well as adding photorespiratory bypasses to reduce carbon loss due to photorespiration. Lastly, work has been done to design a new artificial carbon assimilation cycle by using deep learning and synthetic biology. Synthetic biology approaches can also aid in developing artificial enzyme complexes that could improve photosynthesis in plants. (Summary by Mae Mercado @maemercado.bsky.social) J. Integr. Plant Biol. https://onlinelibrary.wiley.com/doi/10.1111/jipb.13825
Rubisco (Ribulose‐1,5‐bisphosphate carboxylase/oxygenase) is the central enzyme for photosynthesis, This enzyme poorly discriminates between CO2 and O2, which limits its efficiency. To work around this and make carbon assimilation more efficient, scientists have been employing different engineering strategies, several of which are summarized in a new review by Qin et al. One strategy is to alter Rubisco activity, for example by inducing mutations, combining parts of Rubisco from different organisms, engineering other enzymes involved with Rubisco activation and accumulation, and introducing Rubisco genes from lower organisms to crop plants. Another engineering approach is to create carbon-concentrating compartments, for example by using carboxysomes found in cyanobacteria and pyrenoids from eukaryotic algae. Alternatively, CO2 levels at Rubisco can be boosted by introducing the C4 cycle in C3 plants. Scientists are also engineering the Calvin-Benson-Bassham Cycle by overexpressing specific enzymes involved in the cycle, as well as adding photorespiratory bypasses to reduce carbon loss due to photorespiration. Lastly, work has been done to design a new artificial carbon assimilation cycle by using deep learning and synthetic biology. Synthetic biology approaches can also aid in developing artificial enzyme complexes that could improve photosynthesis in plants. (Summary by Mae Mercado @maemercado.bsky.social) J. Integr. Plant Biol. https://onlinelibrary.wiley.com/doi/10.1111/jipb.13825
Review: Geography, altitude, agriculture, and hypoxia
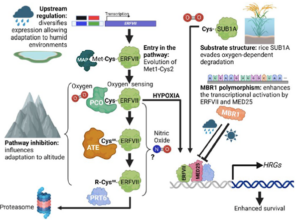 Hypoxia, or reduced oxygen availability, is a double-edged sword: while it disrupts metabolism and can cause cell death, it also plays a vital role in regulating development in animals and plants. With extreme flooding increasing due to climate change, understanding how genetic variation enhances hypoxia tolerance provides valuable strategies for improving crop resilience. This review by Holdsworth et al. examines how geography, altitude, and agriculture have shaped molecular responses to hypoxia, particularly through the PLANT CYSTEINE OXIDASE (PCO) N-degron pathway, and how these responses have evolved across different land plant species over time. Local adaptation plays a key role in acclimatization to humid environments, where flooding, often triggered by intense rainfall, drives acute hypoxic stress. Environmental genome-wide association studies (eGWAS) help identify genes linked to hypoxia tolerance by integrating large-scale genomic and environmental data. Altitude also influences oxygen-sensing adaptations, though no direct molecular mechanism for absolute altitude sensing has been identified. However, adaptation to altitude is at least partially linked to the PCO N-degron pathway. Agriculture has further shaped hypoxia responses, as seen in rice: the FR13A variety uses the Sub1A-1 gene to withstand full submersion by limiting growth and enhancing anaerobic metabolism, while deepwater rice employs SNORKEL1/2 genes to elongate stems and escape partial submersion. These insights drive breeding strategies for flood-resistant crops. Ultimately, understanding the conserved PCO N-degron pathway provides a foundation for future research into plant adaptation to low-oxygen environments, integrating georeferenced genomic data and biochemical findings. This article is part of the Plant Physiology focus issue on Hypoxia and Plants; a recorded webinar on this topic can be found here. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) Plant Physiol. https://doi.org/10.1093/plphys/kiae535
Hypoxia, or reduced oxygen availability, is a double-edged sword: while it disrupts metabolism and can cause cell death, it also plays a vital role in regulating development in animals and plants. With extreme flooding increasing due to climate change, understanding how genetic variation enhances hypoxia tolerance provides valuable strategies for improving crop resilience. This review by Holdsworth et al. examines how geography, altitude, and agriculture have shaped molecular responses to hypoxia, particularly through the PLANT CYSTEINE OXIDASE (PCO) N-degron pathway, and how these responses have evolved across different land plant species over time. Local adaptation plays a key role in acclimatization to humid environments, where flooding, often triggered by intense rainfall, drives acute hypoxic stress. Environmental genome-wide association studies (eGWAS) help identify genes linked to hypoxia tolerance by integrating large-scale genomic and environmental data. Altitude also influences oxygen-sensing adaptations, though no direct molecular mechanism for absolute altitude sensing has been identified. However, adaptation to altitude is at least partially linked to the PCO N-degron pathway. Agriculture has further shaped hypoxia responses, as seen in rice: the FR13A variety uses the Sub1A-1 gene to withstand full submersion by limiting growth and enhancing anaerobic metabolism, while deepwater rice employs SNORKEL1/2 genes to elongate stems and escape partial submersion. These insights drive breeding strategies for flood-resistant crops. Ultimately, understanding the conserved PCO N-degron pathway provides a foundation for future research into plant adaptation to low-oxygen environments, integrating georeferenced genomic data and biochemical findings. This article is part of the Plant Physiology focus issue on Hypoxia and Plants; a recorded webinar on this topic can be found here. (Summary by Elisa De Meo, www.linkedin.com/in/elisa-de-meo-25415a20b) Plant Physiol. https://doi.org/10.1093/plphys/kiae535
Structural insights into PHO1: A key regulator of phosphate translocation in plants
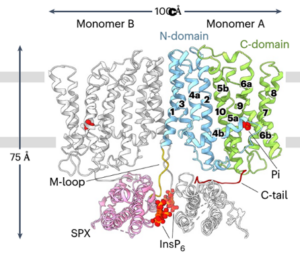 Phosphorus (P) is an essential macronutrient required for plant growth, development, and reproduction. It is primarily absorbed by plant roots in the form of orthophosphate (Pi). The root-to-shoot translocation of Pi depends on a crucial xylem-loading process mediated by PHOSPHATE 1 (PHO1), a Pi efflux transporter expressed in the pericycle and xylem parenchyma cells. Mutations in PHO1 result in severely impaired shoot growth and reduced seed production. Although the physiological role of PHO1 has been recognized for over two decades, its structure and gating mechanism were only recently elucidated. Using a combination of crystallography and modeling tools, Fang and colleagues resolved the Arabidopsis PHO1;H1 (AtPHO1;H1) structure, revealing an N-terminal cytoplasmic SPX domain, a transmembrane EXS domain, and a partially disordered C-terminal tail. The EXS domain facilitates Pi translocation and is gated by the residues Trp719 and Tyr610. The SPX domain binds PP-InsP molecules to regulate transport activity, while the C-terminal tail contributes to dimer formation and PP-InsP binding. These findings provide critical insights for optimizing phosphorus-use efficiency (PUE) in plants, which is essential for reducing dependence on P fertilizers and promoting sustainable, environmentally friendly agriculture. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-024-01895-6
Phosphorus (P) is an essential macronutrient required for plant growth, development, and reproduction. It is primarily absorbed by plant roots in the form of orthophosphate (Pi). The root-to-shoot translocation of Pi depends on a crucial xylem-loading process mediated by PHOSPHATE 1 (PHO1), a Pi efflux transporter expressed in the pericycle and xylem parenchyma cells. Mutations in PHO1 result in severely impaired shoot growth and reduced seed production. Although the physiological role of PHO1 has been recognized for over two decades, its structure and gating mechanism were only recently elucidated. Using a combination of crystallography and modeling tools, Fang and colleagues resolved the Arabidopsis PHO1;H1 (AtPHO1;H1) structure, revealing an N-terminal cytoplasmic SPX domain, a transmembrane EXS domain, and a partially disordered C-terminal tail. The EXS domain facilitates Pi translocation and is gated by the residues Trp719 and Tyr610. The SPX domain binds PP-InsP molecules to regulate transport activity, while the C-terminal tail contributes to dimer formation and PP-InsP binding. These findings provide critical insights for optimizing phosphorus-use efficiency (PUE) in plants, which is essential for reducing dependence on P fertilizers and promoting sustainable, environmentally friendly agriculture. (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-024-01895-6
High-energy requiring pollen grains have specialized mitochondria
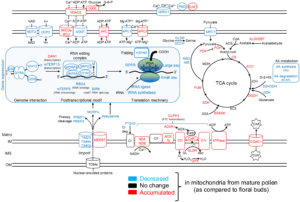 Imagine you’re on a quest to deliver a package, racing against the competition. How do you prepare? Pollen grains and the pollen tubes that they form are essentially package-delivery systems. Their purpose is to deliver genetic information (sperm cell nuclei) to the ovule. Once the task is completed, the pollen cell itself becomes obsolete. A new study by Boussardon et al. investigated the proteome of mature pollen grains which they isolated using the IMTACT protocol: Isolation of Mitochondria Tagged in specific Cell-Type. The authors showed that pollen grain mitochondria switch into a focused energy-generating program, with proteins involved in energy production (TCA cycle and electron transport chain) highly enriched, not unlike a runner carbo-loading before a race. At the same time, proteins involved in transcription and translation are strongly depleted, and the mitochondrial genome itself is actively degraded during maturation; in essence the mitochondria terminally differentiate and channel their resources into their delivery task. This observation is fascinating in light of the fact that the progeny of sexual reproduction inherit their mitochondria solely from the egg-donating parent, and not the sperm-donating parent, no pollen grain mitochondria need to be viable for transfer into the zygote. (Summary by Mary Williams @PlantTeaching.bsky.social ) Curr. Biol. 10.1016/j.cub.2024.12.037
Imagine you’re on a quest to deliver a package, racing against the competition. How do you prepare? Pollen grains and the pollen tubes that they form are essentially package-delivery systems. Their purpose is to deliver genetic information (sperm cell nuclei) to the ovule. Once the task is completed, the pollen cell itself becomes obsolete. A new study by Boussardon et al. investigated the proteome of mature pollen grains which they isolated using the IMTACT protocol: Isolation of Mitochondria Tagged in specific Cell-Type. The authors showed that pollen grain mitochondria switch into a focused energy-generating program, with proteins involved in energy production (TCA cycle and electron transport chain) highly enriched, not unlike a runner carbo-loading before a race. At the same time, proteins involved in transcription and translation are strongly depleted, and the mitochondrial genome itself is actively degraded during maturation; in essence the mitochondria terminally differentiate and channel their resources into their delivery task. This observation is fascinating in light of the fact that the progeny of sexual reproduction inherit their mitochondria solely from the egg-donating parent, and not the sperm-donating parent, no pollen grain mitochondria need to be viable for transfer into the zygote. (Summary by Mary Williams @PlantTeaching.bsky.social ) Curr. Biol. 10.1016/j.cub.2024.12.037
You’ve gotta starch somewhere: Evidence for an alternative starch granule initiation pathway
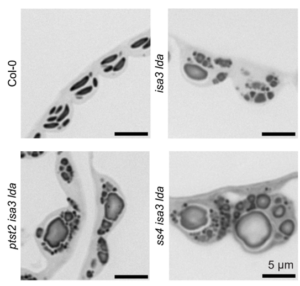
Starch is the major storage carbohydrate in plants, and in Arabidopsis leaves it forms granules in the chloroplasts to supply energy during the night when photosynthesis is inactive. These semicrystalline granules are made of glucose chains, which can be mostly unbranched (amylose) or highly branched (amylopectin). The formation of new starch granules (initiation) is achieved by building on short glucose chains called malto-oligosaccharide (MOS) primers. A key step in this process is the delivery of a glucosyltransferase STARCH SYNTHASE 4 (SS4) to the new granule by PROTEIN TARGETING TO STARCH 2 (PTST2). However, new evidence from Heutinck et al. supports an alternative route for starch granule initiation. In an Arabidopsis mutant deficient in two debranching enzymes, branched MOS in the chloroplast accumulate to levels ten times higher than in the wild type. Additional granules were also formed in these chloroplasts, which were smaller, differently shaped, and not reliant on SS4 or PTST2 to be initiated. While additional research is needed to elucidate the mechanism, the possibility of a new pathway for starch granule initiation provides huge potential for better understanding plant carbon metabolism and hence crop improvement. (Summary by Ciara O’Brien) Plant Physiol. 10.1093/plphys/kiaf002
Prion-like domains of sensory HSFs remember heat
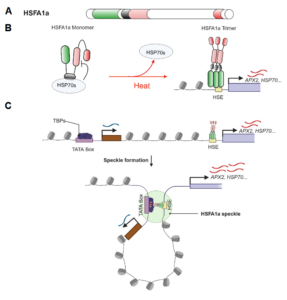 The heat shock response, a rapid transcriptional response to heat, was first observed nearly 60 years ago, and has long been a paradigm for understanding gene responses to exogenous cues. The family of genes encoding heat shock factors (HSFs) is greatly expanded in plants. These HSFs serve as key regulators of the heat shock response, both initiating short-term transcriptional changes, but also serving as a molecular memory that activates heat memory genes after an earlier “priming” stress. A new study by Peng et al. examined how conformational changes in HSF protein structure contributes to both short term and memory responses to heat shock. The authors identified a transcriptional cascade, led by rapid activation HSFA1 by protein conformational changes. Within moments of heat shock, HSFA1 factors form speckles in the nucleus, suggesting that they act as heat sensors, and eliciting a very rapid transcriptional response. HSFA1 factors contain two Prion-related Domains (PrDs), which the authors showed are critical for their thermal responsiveness. PrDs are intrinsically disordered domains that are often found in proteins that undergo liquid phase separation. One of these, PrD1, is responsible for sequestering the HSFA1 proteins during ambient temperatures and maintaining them in an inactive state. The other, PrD2, is required for heat activation and thermal memory, through the formation of DNA loops that connect heat-responsive promoters with enhancer domains. This work builds on the growing recognition of the importance of intrinsically disordered domains and liquid phase separation in modulating rapid cellular responses. (Summary by Mary Williams @PlantTeaching.bsky.social) Mol Plant 10.1016/j.molp.2025.01.007
The heat shock response, a rapid transcriptional response to heat, was first observed nearly 60 years ago, and has long been a paradigm for understanding gene responses to exogenous cues. The family of genes encoding heat shock factors (HSFs) is greatly expanded in plants. These HSFs serve as key regulators of the heat shock response, both initiating short-term transcriptional changes, but also serving as a molecular memory that activates heat memory genes after an earlier “priming” stress. A new study by Peng et al. examined how conformational changes in HSF protein structure contributes to both short term and memory responses to heat shock. The authors identified a transcriptional cascade, led by rapid activation HSFA1 by protein conformational changes. Within moments of heat shock, HSFA1 factors form speckles in the nucleus, suggesting that they act as heat sensors, and eliciting a very rapid transcriptional response. HSFA1 factors contain two Prion-related Domains (PrDs), which the authors showed are critical for their thermal responsiveness. PrDs are intrinsically disordered domains that are often found in proteins that undergo liquid phase separation. One of these, PrD1, is responsible for sequestering the HSFA1 proteins during ambient temperatures and maintaining them in an inactive state. The other, PrD2, is required for heat activation and thermal memory, through the formation of DNA loops that connect heat-responsive promoters with enhancer domains. This work builds on the growing recognition of the importance of intrinsically disordered domains and liquid phase separation in modulating rapid cellular responses. (Summary by Mary Williams @PlantTeaching.bsky.social) Mol Plant 10.1016/j.molp.2025.01.007
How a model C4 plant, Setaria viridis, copes with prolonged heat
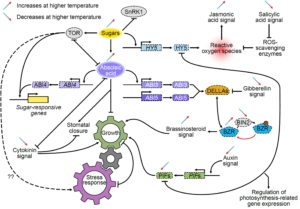 Due to anthropomorphic global warming, 2024 was the first year during which the global mean temperature was more than 1.5° above pre-industrial levels. Clearly, understanding how high temperatures affect plant physiology is an urgent priority. In this new study, Zhang et al. did a multi-parameter study of the model C4 plant Setaria viridis. They grew two populations for two weeks at 28°, and then for the next two weeks one group was grown at 42° whilst the other remained at 28°. After this, the authors measured photosynthesis parameters, and carried out metabolomic, transcriptomic, and proteomic studies, leading to several key findings. As expected, the heat-grown plants were significantly smaller than their counterparts. Surprisingly, their photosynthetic efficiency was hardly altered. The heat-grown plants had much higher levels of hexoses, which might be a protective mechanism to retain water. The authors also found increased levels of ABA and amino-acid conjugates of IAA, which may contribute to the smaller size of the plants. There’s a LOT of interesting data, so have a look. Your favorite gene or metabolite is likely to appear in this useful and comprehensive study. (Summary by Mary Williams @plantteaching.bsky.social) Plant Cell https://doi.org/10.1093/plcell/koaf005
Due to anthropomorphic global warming, 2024 was the first year during which the global mean temperature was more than 1.5° above pre-industrial levels. Clearly, understanding how high temperatures affect plant physiology is an urgent priority. In this new study, Zhang et al. did a multi-parameter study of the model C4 plant Setaria viridis. They grew two populations for two weeks at 28°, and then for the next two weeks one group was grown at 42° whilst the other remained at 28°. After this, the authors measured photosynthesis parameters, and carried out metabolomic, transcriptomic, and proteomic studies, leading to several key findings. As expected, the heat-grown plants were significantly smaller than their counterparts. Surprisingly, their photosynthetic efficiency was hardly altered. The heat-grown plants had much higher levels of hexoses, which might be a protective mechanism to retain water. The authors also found increased levels of ABA and amino-acid conjugates of IAA, which may contribute to the smaller size of the plants. There’s a LOT of interesting data, so have a look. Your favorite gene or metabolite is likely to appear in this useful and comprehensive study. (Summary by Mary Williams @plantteaching.bsky.social) Plant Cell https://doi.org/10.1093/plcell/koaf005
Streamlining the immune system: How plants adapt to reduced pathogen pressure
 Although plants are sessile organisms unable to escape pathogen invasions, they are well equipped with defense mechanisms. Cell surface-localized pattern-recognition receptors (PRRs) detect extracellular signals and initiate pattern-triggered immunity (PTI), while nucleotide-binding leucine-rich repeat receptors (NLRs) sense intracellular signals and trigger effector-triggered immunity (ETI). These two layers of immune signaling were long considered distinct pathways but are now recognized as interdependent—essentially functioning as one. While numerous studies have explored the expansion of PRRs and NLRs during plant evolution, particularly during the transition from aquatic to terrestrial environments, the opposite scenario remains less examined: how plants with specialized lifestyles—such as mangrove, desert, or aquatic species—adapt to reduced pathogen pressure. Li and colleagues reassessed 808 angiosperm genomes and found that plants with specialized lifestyles and habitats (SLHs) exhibit significant reductions in PRR and NLR genes due to increased gene loss and reduced gene duplication. Evolutionary association studies further identified key immune regulatory modules, including the well-known EDS1, alongside novel components. These newly identified modules and annotated immune-related genes from 808 angiosperm species offer valuable insights into plant immunity and can be explored through the publicly available database, AirDB (https://efg.nju.edu.cn/AirDB/). (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-024-01901-x
Although plants are sessile organisms unable to escape pathogen invasions, they are well equipped with defense mechanisms. Cell surface-localized pattern-recognition receptors (PRRs) detect extracellular signals and initiate pattern-triggered immunity (PTI), while nucleotide-binding leucine-rich repeat receptors (NLRs) sense intracellular signals and trigger effector-triggered immunity (ETI). These two layers of immune signaling were long considered distinct pathways but are now recognized as interdependent—essentially functioning as one. While numerous studies have explored the expansion of PRRs and NLRs during plant evolution, particularly during the transition from aquatic to terrestrial environments, the opposite scenario remains less examined: how plants with specialized lifestyles—such as mangrove, desert, or aquatic species—adapt to reduced pathogen pressure. Li and colleagues reassessed 808 angiosperm genomes and found that plants with specialized lifestyles and habitats (SLHs) exhibit significant reductions in PRR and NLR genes due to increased gene loss and reduced gene duplication. Evolutionary association studies further identified key immune regulatory modules, including the well-known EDS1, alongside novel components. These newly identified modules and annotated immune-related genes from 808 angiosperm species offer valuable insights into plant immunity and can be explored through the publicly available database, AirDB (https://efg.nju.edu.cn/AirDB/). (Summary by Ching Chan @ntnuchanlab) Nature Plants 10.1038/s41477-024-01901-x
Double attack! Herbivore insects feed on plants and silence their genes
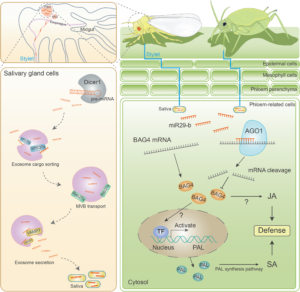 In the natural environment, plants are constantly attacked by animals. Plant immunity is regulated by a network of genes and hormones including salicylic acid (SA) and jasmonic acid (JA). A recent study suggests that herbivore insects threaten plants with concerns more than the bite. Han et al., found that the phloem-feeding whitefly (Bemisa tabaci) transfers miRNA BtmiR29-b to tobacco phloem sap during feeding. The miRNA suppresses plant defense by silencing the defense gene BAG4 and inhibits SA and JA accumulations in the host. Searching through the miRNA database miRBase, the authors found that miR29-b exists in many other insect species of various orders including Hemiptera, Coleoptera, Hymenoptera, Orthoptera, and Blattaria. Besides whitefly, miR29-b was experimentally detected in the salivary gland and saliva of aphid (Myzus persicae), which could transfer the miRNA to tobacco phloem sap during feeding. In plants, BAG4 homologs exist in various species of different families including Solanaceae, Brassicaceae, Cucurbitaceae, Euphorbiaceae, Leguminosae, and Malvaceae. Despite the different target sites, the targeting of the homologous BAG4 transcripts by miR29-b could be predicted. The transfer of BtmiR29-b from whiteflies to Arabidopsis, cotton, and tomato was also experimentally validated. The study demonstrates cross-kingdom miRNA-mediated gene silencing and suggests the conserved mechanism in different species. The findings motivate future studies in other cross-kingdom gene regulations and provide insights into pest control strategies. (Summary by Yee-Shan Ku @YeeShanKu1) Molecular Plant 10.1016/j.molp.2025.01.001
In the natural environment, plants are constantly attacked by animals. Plant immunity is regulated by a network of genes and hormones including salicylic acid (SA) and jasmonic acid (JA). A recent study suggests that herbivore insects threaten plants with concerns more than the bite. Han et al., found that the phloem-feeding whitefly (Bemisa tabaci) transfers miRNA BtmiR29-b to tobacco phloem sap during feeding. The miRNA suppresses plant defense by silencing the defense gene BAG4 and inhibits SA and JA accumulations in the host. Searching through the miRNA database miRBase, the authors found that miR29-b exists in many other insect species of various orders including Hemiptera, Coleoptera, Hymenoptera, Orthoptera, and Blattaria. Besides whitefly, miR29-b was experimentally detected in the salivary gland and saliva of aphid (Myzus persicae), which could transfer the miRNA to tobacco phloem sap during feeding. In plants, BAG4 homologs exist in various species of different families including Solanaceae, Brassicaceae, Cucurbitaceae, Euphorbiaceae, Leguminosae, and Malvaceae. Despite the different target sites, the targeting of the homologous BAG4 transcripts by miR29-b could be predicted. The transfer of BtmiR29-b from whiteflies to Arabidopsis, cotton, and tomato was also experimentally validated. The study demonstrates cross-kingdom miRNA-mediated gene silencing and suggests the conserved mechanism in different species. The findings motivate future studies in other cross-kingdom gene regulations and provide insights into pest control strategies. (Summary by Yee-Shan Ku @YeeShanKu1) Molecular Plant 10.1016/j.molp.2025.01.001