Plant Science Research Weekly: February 16, 2024
Review: Integrating cellular electron microscopy with multimodal data to explore biology across space and time
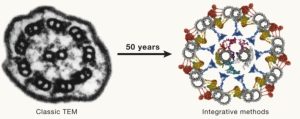 Fifty years ago (1974), Albert Claude, Christian de Duve, and George Palade were awarded the Nobel Prize for their discoveries on the structural and functional organization of the cell, which Claude eloquently framed by writing, “We have entered the cell, the mansion of our birth, and started the inventory of our acquired wealth.” In the subsequent fifty years, amazing new tools and technologies have greatly enhanced our understanding of our cellular inventory. McCafferty et al. have skillfully crafted a comprehensive overview of these methods, beautifully weaving together how they complement each other to provide extraordinary insights into cell structures and compositions in space and time. I particularly enjoyed how the authors melded multiple imaging technologies together along with computational and modeling approaches. As one of several examples, they show using Chlamydomonas how fluorescent microscopy can be combined with ultrastructure expansion microscopy, soft X-ray tomography, cross-linking and co-expression mass spectrometry, single-particle analysis, alpha fold structure prediction, proximity labeling and molecular dynamics modeling. This is a fascinating and inspiring article that makes me eager to see where the next 50 years will take us. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2024.01.005
Fifty years ago (1974), Albert Claude, Christian de Duve, and George Palade were awarded the Nobel Prize for their discoveries on the structural and functional organization of the cell, which Claude eloquently framed by writing, “We have entered the cell, the mansion of our birth, and started the inventory of our acquired wealth.” In the subsequent fifty years, amazing new tools and technologies have greatly enhanced our understanding of our cellular inventory. McCafferty et al. have skillfully crafted a comprehensive overview of these methods, beautifully weaving together how they complement each other to provide extraordinary insights into cell structures and compositions in space and time. I particularly enjoyed how the authors melded multiple imaging technologies together along with computational and modeling approaches. As one of several examples, they show using Chlamydomonas how fluorescent microscopy can be combined with ultrastructure expansion microscopy, soft X-ray tomography, cross-linking and co-expression mass spectrometry, single-particle analysis, alpha fold structure prediction, proximity labeling and molecular dynamics modeling. This is a fascinating and inspiring article that makes me eager to see where the next 50 years will take us. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2024.01.005
Review. Mycorrhizal symbiosis: Genomics, ecology, and agricultural application
 This outstanding Tansley review by Martin and van der Heijden spans the scale of research on mycorrhizal symbiosis from molecules to ecosystems, and spans time from the earliest encroachment of plants and fungi onto land to the future applications of our understanding. This very comprehensive review is a great resource to anyone with an interest in mycorrhizal symbiosis. I particularly appreciate their objective approach to addressing some of the potentially overhyped topics. For example, the idea of plants communicating and displaying altruistic behaviors through common mycorrhizal networks is intriguing, but the evidence for such effects is small. The authors also look at the potential for improving plant productivity through mycorrhizal inoculation. Although some studies have shown growth enhancement, the range of responses is huge and highly dependent on many factors that are difficult to identify and control. Mycorrhizal symbiosis impacts on and by climate change are also discussed, as are the many questions that require further research. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19541
This outstanding Tansley review by Martin and van der Heijden spans the scale of research on mycorrhizal symbiosis from molecules to ecosystems, and spans time from the earliest encroachment of plants and fungi onto land to the future applications of our understanding. This very comprehensive review is a great resource to anyone with an interest in mycorrhizal symbiosis. I particularly appreciate their objective approach to addressing some of the potentially overhyped topics. For example, the idea of plants communicating and displaying altruistic behaviors through common mycorrhizal networks is intriguing, but the evidence for such effects is small. The authors also look at the potential for improving plant productivity through mycorrhizal inoculation. Although some studies have shown growth enhancement, the range of responses is huge and highly dependent on many factors that are difficult to identify and control. Mycorrhizal symbiosis impacts on and by climate change are also discussed, as are the many questions that require further research. (Summary by Mary Williams @PlantTeaching) New Phytol. 10.1111/nph.19541
Oxygen supply dictates growth and metabolism in young leaves
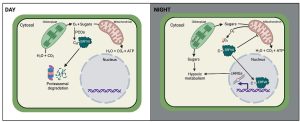 When thinking of hypoxia or low oxygen in plants, the image that often comes to mind is one of flooding stress. However, it’s not just plants exposed to excess of water that face hypoxia. Even in growing plants, hypoxia sensing and the existence of hypoxic niches play a vital role in their development. Young, actively growing leaves have different metabolic needs compared to mature leaves, characterized by high respiration rates and thus high oxygen consumption. Yet a functional connection between internal oxygen sensing and a metabolic shift in developing plants is unexplored. In this intriguing study, Triozzi et al. uncovered the existence of cyclic hypoxia in young leaves. The authors found that the expression of hypoxia-responsive genes (HRGs) exhibits diurnal fluctuations, with peak expression at night. Using a combination of genetic and pharmacological approaches and transcriptional reporters, in combination with oxygen-modified atmosphere treatments, they demonstrated that diurnal variations in HRGs occur independently of light and clock inputs and are actually regulated by oxygen levels. Through the measurement of internal oxygen levels, the authors reveal that young leaves, and not old leaves, experience a drop in oxygen during the night. This nocturnal oxygen dip and the associated hypoxia signaling cascade, which is ERFVII-dependent, causes a shift in metabolism from aerobic to hypoxic metabolism, which is important to modulate leaf growth at at different times of the day. (Summary by Thomas Depaepe @thdpaepe) Mol. Plant 10.1016/j.molp.2024.01.006
When thinking of hypoxia or low oxygen in plants, the image that often comes to mind is one of flooding stress. However, it’s not just plants exposed to excess of water that face hypoxia. Even in growing plants, hypoxia sensing and the existence of hypoxic niches play a vital role in their development. Young, actively growing leaves have different metabolic needs compared to mature leaves, characterized by high respiration rates and thus high oxygen consumption. Yet a functional connection between internal oxygen sensing and a metabolic shift in developing plants is unexplored. In this intriguing study, Triozzi et al. uncovered the existence of cyclic hypoxia in young leaves. The authors found that the expression of hypoxia-responsive genes (HRGs) exhibits diurnal fluctuations, with peak expression at night. Using a combination of genetic and pharmacological approaches and transcriptional reporters, in combination with oxygen-modified atmosphere treatments, they demonstrated that diurnal variations in HRGs occur independently of light and clock inputs and are actually regulated by oxygen levels. Through the measurement of internal oxygen levels, the authors reveal that young leaves, and not old leaves, experience a drop in oxygen during the night. This nocturnal oxygen dip and the associated hypoxia signaling cascade, which is ERFVII-dependent, causes a shift in metabolism from aerobic to hypoxic metabolism, which is important to modulate leaf growth at at different times of the day. (Summary by Thomas Depaepe @thdpaepe) Mol. Plant 10.1016/j.molp.2024.01.006
Phase separation determining meiotic interference-sensitive meiotic crossover formation
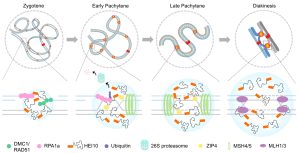 In sexual reproduction, meiosis creates new combinations between homologs and diversifies the genetic information among gametes and progenies. Most meiotic crossovers (COs) are mediated by a group of ZMM factors, including HUMAN ENHANCER of INVASION-10 (HEI10), which is suggested to concentrate at CO sites to form larger foci for crossover interference. However, how HEI10 is recruited to the CO site is still unclear. Here, Wang et al. showed Arabidopsis HEI10 undergoes liquid-liquid phase separation (LLPS) and its condensation depends on its residue Ser70. The HEI10 promoted the ubiquitination-dependent degradation of the interacting REPLICATION PROTEIN A (RPA1a) in the LLPS of CO sites through the early pachytene phase. This study also provides clues about how HEI10 controls class I meiotic crossover formation and can be relevant in most eukaryotes. (Summary by Yueh Cho @YuehCho1984) PNAS. 10.1073/pnas.2310542120.
In sexual reproduction, meiosis creates new combinations between homologs and diversifies the genetic information among gametes and progenies. Most meiotic crossovers (COs) are mediated by a group of ZMM factors, including HUMAN ENHANCER of INVASION-10 (HEI10), which is suggested to concentrate at CO sites to form larger foci for crossover interference. However, how HEI10 is recruited to the CO site is still unclear. Here, Wang et al. showed Arabidopsis HEI10 undergoes liquid-liquid phase separation (LLPS) and its condensation depends on its residue Ser70. The HEI10 promoted the ubiquitination-dependent degradation of the interacting REPLICATION PROTEIN A (RPA1a) in the LLPS of CO sites through the early pachytene phase. This study also provides clues about how HEI10 controls class I meiotic crossover formation and can be relevant in most eukaryotes. (Summary by Yueh Cho @YuehCho1984) PNAS. 10.1073/pnas.2310542120.
Signaling secrets: FERONIA´s dynamic interplay with interactors at the cell surface
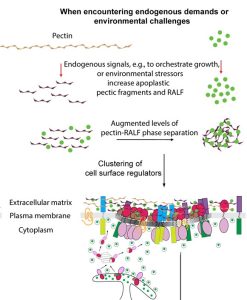 Named after the Etruscan goddess of fertility, the receptor-like kinase FERONIA (FER) is involved in more than fertility. A recent paper by Liu, Yeh et al. sheds light on the intricate signaling processes taking place on the cell surface in response to environmental signals regulating growth and development of plant cells. From previous studies, we know that FER can bind the cell wall polysaccharide pectin and interact with its peptide ligand, rapid alkalinization factor1 (RALF1). In this paper, the current signaling model is expanded. What is striking here is that the RALF1-FER interaction involves pectin but also LLG1 (LORELEI-LIKE glycosylphosphatidylinositol-anchored protein 1), a co-receptor of FER, guiding FER to its functional location. The molecular condensates formed by phase separation of RALF1 and pectin in the apoplastic space function as surface sensors for stress. The authors demonstrate that upon binding of the peptide ligand RALF1 at the plasma membrane, FER and LLG1 are internalized, concentrating in a nanodomain at the membrane. Interestingly, RALF1 itself does not get taken up into the cell, remaining in the apoplast. This interaction represents the initial step in a signal cascade in order to trigger downstream responses. The authors note that these condensates are further stimulated by environmental changes, activating a variety of downstream responses, including those associated with stresses such as salt and heat. This study highlights the connection between extracellular processes and intracellular responses implemented initially by upregulated endocytosis and expands our understanding of the diverse biological role of FER. (Summary by Ann-Kathrin Rößling @AK_Roessling) Cell 10.1016/j.cell.2023.11.038
Named after the Etruscan goddess of fertility, the receptor-like kinase FERONIA (FER) is involved in more than fertility. A recent paper by Liu, Yeh et al. sheds light on the intricate signaling processes taking place on the cell surface in response to environmental signals regulating growth and development of plant cells. From previous studies, we know that FER can bind the cell wall polysaccharide pectin and interact with its peptide ligand, rapid alkalinization factor1 (RALF1). In this paper, the current signaling model is expanded. What is striking here is that the RALF1-FER interaction involves pectin but also LLG1 (LORELEI-LIKE glycosylphosphatidylinositol-anchored protein 1), a co-receptor of FER, guiding FER to its functional location. The molecular condensates formed by phase separation of RALF1 and pectin in the apoplastic space function as surface sensors for stress. The authors demonstrate that upon binding of the peptide ligand RALF1 at the plasma membrane, FER and LLG1 are internalized, concentrating in a nanodomain at the membrane. Interestingly, RALF1 itself does not get taken up into the cell, remaining in the apoplast. This interaction represents the initial step in a signal cascade in order to trigger downstream responses. The authors note that these condensates are further stimulated by environmental changes, activating a variety of downstream responses, including those associated with stresses such as salt and heat. This study highlights the connection between extracellular processes and intracellular responses implemented initially by upregulated endocytosis and expands our understanding of the diverse biological role of FER. (Summary by Ann-Kathrin Rößling @AK_Roessling) Cell 10.1016/j.cell.2023.11.038
RAF-like protein kinases mediate a deeply conserved, rapid auxin response
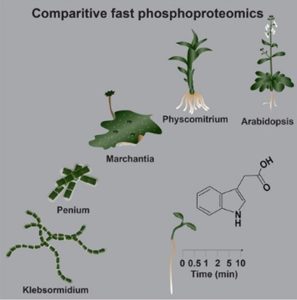 The definition of a phytohormone has not always been clear, though most plant biologists agree that phytohormones can be defined as endogenous molecules capable of triggering signaling cascades and associated responses by binding to specific receptors. They have been extensively studied in flowering plants, but in the last decade we witnessed a significant shift in research focus towards algae, bryophytes, and other land plants. This perspective provided novel insights into hormone evolution and their function. While metabolomics and genomics have proven that the synthesis of certain hormones in various algal sister groups of plants does occur, their role as true signals with a signaling machinery remains contested. A recent study by Kuhn et al. revealed that, like in Arabidopsis, auxin induces a global phosphorylation response in two species of streptophyte algae. The authors demonstrated that auxin induces membrane polarization and cytoplasmic streaming in Klebsormidium, classified as so-called ‘fast’ auxin responses. Phosphoproteomics further revealed a rapid, dynamic and auxin-specific phosphorylation response to be conserved in several lineages. In silico analysis then identified a group of B4 RAF-like kinases as central mediators of the auxin phosphoresponse. Moreover, loss-of-function mutant phenotypes and transcriptome analyses confirmed their crucial role in rapid auxin signaling, influencing overall growth and development. Despite the central involvement of these RAF kinases, the perception mechanism of auxin in algae remains unknown and will be an exciting topic for further investigation. (Summary by Thomas Depaepe @thdpaepe) 10.1016/j.cell.2023.11.021
The definition of a phytohormone has not always been clear, though most plant biologists agree that phytohormones can be defined as endogenous molecules capable of triggering signaling cascades and associated responses by binding to specific receptors. They have been extensively studied in flowering plants, but in the last decade we witnessed a significant shift in research focus towards algae, bryophytes, and other land plants. This perspective provided novel insights into hormone evolution and their function. While metabolomics and genomics have proven that the synthesis of certain hormones in various algal sister groups of plants does occur, their role as true signals with a signaling machinery remains contested. A recent study by Kuhn et al. revealed that, like in Arabidopsis, auxin induces a global phosphorylation response in two species of streptophyte algae. The authors demonstrated that auxin induces membrane polarization and cytoplasmic streaming in Klebsormidium, classified as so-called ‘fast’ auxin responses. Phosphoproteomics further revealed a rapid, dynamic and auxin-specific phosphorylation response to be conserved in several lineages. In silico analysis then identified a group of B4 RAF-like kinases as central mediators of the auxin phosphoresponse. Moreover, loss-of-function mutant phenotypes and transcriptome analyses confirmed their crucial role in rapid auxin signaling, influencing overall growth and development. Despite the central involvement of these RAF kinases, the perception mechanism of auxin in algae remains unknown and will be an exciting topic for further investigation. (Summary by Thomas Depaepe @thdpaepe) 10.1016/j.cell.2023.11.021
Sealing the deal to graft healing: PAT1 as a novel regulator of graft formation
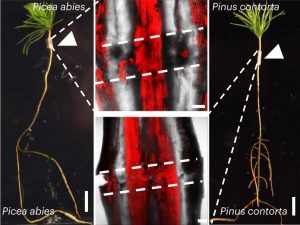 Grafting is a way to combine the desirable properties of two plants by joining the scions and rootstocks from different plants. While frequently practiced, little is known about how grafting within and between coniferous species works. Most pertinently, grafting success relies on how efficiently connected vasculatures can heal at the graft junction. Precisely, this is what Feng et al. have demonstrated in their article. To observe graft healing dynamics, the authors developed a new micrografting technique that allows successful grafting between young tissues of divergent conifer species, which was previously inhibited by graft incompatibility. By performing RNA sequencing on intact and grafted Norway Spruce (Picea abies), the authors were able to identify an upregulated gene, PHYTOCHROME A SIGNAL TRANSDUCTION 1 (PAT1), common to Arabidopsis and P. abies, using transcriptomic data of Arabidopsis during graft formation. Congruent to this finding, both complementation and overexpression of PaPAT1 in Arabidopsis facilitated graft attachment and callus formation, respectively, whereas loss-of-function PAT mutants are defective in phloem reattachment. These findings reaffirmed the relevance of PAT1 and its orthologs in regulating graft healing in seed plants. (Summary by Marvin Jin @MarvinJYS) Nat. Plants 10.1038/s41477-023-01568-w
Grafting is a way to combine the desirable properties of two plants by joining the scions and rootstocks from different plants. While frequently practiced, little is known about how grafting within and between coniferous species works. Most pertinently, grafting success relies on how efficiently connected vasculatures can heal at the graft junction. Precisely, this is what Feng et al. have demonstrated in their article. To observe graft healing dynamics, the authors developed a new micrografting technique that allows successful grafting between young tissues of divergent conifer species, which was previously inhibited by graft incompatibility. By performing RNA sequencing on intact and grafted Norway Spruce (Picea abies), the authors were able to identify an upregulated gene, PHYTOCHROME A SIGNAL TRANSDUCTION 1 (PAT1), common to Arabidopsis and P. abies, using transcriptomic data of Arabidopsis during graft formation. Congruent to this finding, both complementation and overexpression of PaPAT1 in Arabidopsis facilitated graft attachment and callus formation, respectively, whereas loss-of-function PAT mutants are defective in phloem reattachment. These findings reaffirmed the relevance of PAT1 and its orthologs in regulating graft healing in seed plants. (Summary by Marvin Jin @MarvinJYS) Nat. Plants 10.1038/s41477-023-01568-w
Contribution of synthetic auxin conjugates to clonal propagation of woody species
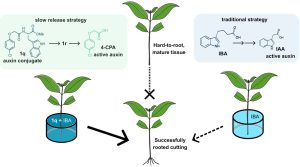 From food and energy to paper and timber, many industries rely on clonal propagation of tree species. Successful propagation of cuttings depends on adventitious rooting ability, which is known to decline with tree age and to be naturally low in many species of interest. Propagators therefore employ naturally occurring auxins, e.g. indole-3-butyric acid (IBA), which has limited positive effect and is often phytotoxic. Roth et al. tested the effect of synthetic auxin conjugates on the rooting ability of Eucalyptus cuttings. When applied with IBA, the 4-chlorophenoxyacetic acid (4-CPA) conjugate 1q particularly enhanced rooting success. Upon penetration into the cutting, 1q is transformed into 1r, which acts a slow-release source of the active synthetic auxin 4-CPA. Then, 1r is cleaved by ILL/ILR1 amido-hydrolases to release 4-CPA. Once released, 4-CPA, which evades the main auxin degradation pathway, can be transported by canonical auxin influx carriers, bind auxin receptors, and trigger adventitious rooting. When applied to other hard-to-root species, 1q often also enhanced their rooting success, even in older individuals. These results show that novel strategies based on slow release of improved auxin formulations can help overcome major roadblocks in clonal tree propagation, which is both key for industry and a crucial tool in reforestation. (Summary by John Vilasboa @vilasjohn) Nature Biotechnol.10.1038/s41587-023-02065-3
From food and energy to paper and timber, many industries rely on clonal propagation of tree species. Successful propagation of cuttings depends on adventitious rooting ability, which is known to decline with tree age and to be naturally low in many species of interest. Propagators therefore employ naturally occurring auxins, e.g. indole-3-butyric acid (IBA), which has limited positive effect and is often phytotoxic. Roth et al. tested the effect of synthetic auxin conjugates on the rooting ability of Eucalyptus cuttings. When applied with IBA, the 4-chlorophenoxyacetic acid (4-CPA) conjugate 1q particularly enhanced rooting success. Upon penetration into the cutting, 1q is transformed into 1r, which acts a slow-release source of the active synthetic auxin 4-CPA. Then, 1r is cleaved by ILL/ILR1 amido-hydrolases to release 4-CPA. Once released, 4-CPA, which evades the main auxin degradation pathway, can be transported by canonical auxin influx carriers, bind auxin receptors, and trigger adventitious rooting. When applied to other hard-to-root species, 1q often also enhanced their rooting success, even in older individuals. These results show that novel strategies based on slow release of improved auxin formulations can help overcome major roadblocks in clonal tree propagation, which is both key for industry and a crucial tool in reforestation. (Summary by John Vilasboa @vilasjohn) Nature Biotechnol.10.1038/s41587-023-02065-3
Discovering and characterizing a protein involved in endosperm starch formation in rice
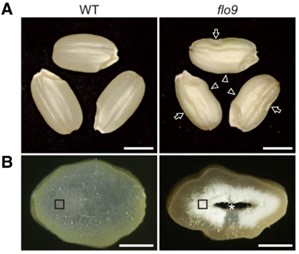 Rice is a globally important crop, with most of its calorific component coming from starch. Despite its importance, we do not fully understand the mechanism of starch formation in the rice endosperm. To discover novel factors involved in this process, Yan et al. performed a forward genetics screen and identified a FLOURY ENDOSPERM 9 (flo9) mutant that had a 32% reduction in starch accumulation. Genetic mapping revealed that flo9 plants had a point mutation in LIKE EARLY STARVATION1 (LESV1) which leads to a premature stop codon and a severely truncated LESV1 protein. LESV1 is a non-enzymatic protein, so the authors investigated whether it interacted with the starch debranching enzyme ISOAMYLASE (ISA1), as isa1 mutants are phenotypically similar to flo9 plants. Yeast 2-hybrid and luciferase complementation assays indeed demonstrated a physical interaction between LESV1 and ISA1. The authors then tested whether the flo9 mutation affects ISA1 localization. Expressing ISA1-GFP in wild type rice protoplasts caused a proportion of ISA1-GFP to be localized to starch granules. Whereas, when repeated with flo9 protoplasts, much less ISA1-GFP localised to starch granules. Hence LESV1 interacts with and targets ISA1 to starch granules and this discovery greatly enhances our understanding of rice endosperm starch formation. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae006
Rice is a globally important crop, with most of its calorific component coming from starch. Despite its importance, we do not fully understand the mechanism of starch formation in the rice endosperm. To discover novel factors involved in this process, Yan et al. performed a forward genetics screen and identified a FLOURY ENDOSPERM 9 (flo9) mutant that had a 32% reduction in starch accumulation. Genetic mapping revealed that flo9 plants had a point mutation in LIKE EARLY STARVATION1 (LESV1) which leads to a premature stop codon and a severely truncated LESV1 protein. LESV1 is a non-enzymatic protein, so the authors investigated whether it interacted with the starch debranching enzyme ISOAMYLASE (ISA1), as isa1 mutants are phenotypically similar to flo9 plants. Yeast 2-hybrid and luciferase complementation assays indeed demonstrated a physical interaction between LESV1 and ISA1. The authors then tested whether the flo9 mutation affects ISA1 localization. Expressing ISA1-GFP in wild type rice protoplasts caused a proportion of ISA1-GFP to be localized to starch granules. Whereas, when repeated with flo9 protoplasts, much less ISA1-GFP localised to starch granules. Hence LESV1 interacts with and targets ISA1 to starch granules and this discovery greatly enhances our understanding of rice endosperm starch formation. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koae006
Patterns of evolution: Dissecting the history of pattern recognition receptors from development to immunity
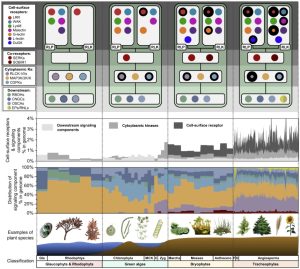 Plants use a suite of cell-surface receptors as signal transducers in both development and immunity. In this study, Ngou et al. utilized computational and functional approaches to examine the evolutionary origin of a subclass of plant cell-surface receptors called pattern recognition receptors (PRRs), as well as their known downstream signalling components. Using 350 genome sequences spanning red algae, green algae, and land plant lineages, they compared the two classes of surface-receptors: receptor-like kinases (RLKs) and receptor-like proteins (RLPs). The group found that RLPs emerged earlier than RLKs, and that the LRR-RLKs and LRR-RLPs showed notable expansions throughout different plant lineages – likely due to their involvement in pathogen detection. Focusing on the largest PRR family, the LRR-RLPs, the authors found a key conserved domain architecture: an island domain (ID) + 4 LRR motifs. Interestingly, they also noticed that the ID + 4 LRRs is also present in the LRR-RLK subgroup Xb, which includes several receptors involved in developmental processes. By performing several domain swapping experiments, they identified regions in the LRR-RLP and LRR-RLK-Xb families responsible for both immune and development pathway activation. The authors propose a persuasive model of “modular evolution,” where specific domains of cell-surface receptors evolve to recognize diverse ligands while other domains remain conserved to preserve distinct downstream signalling pathways. (Summary by Tamar Av-Shalom) Nat Commun. 10.1038/s41467-023-44408-3
Plants use a suite of cell-surface receptors as signal transducers in both development and immunity. In this study, Ngou et al. utilized computational and functional approaches to examine the evolutionary origin of a subclass of plant cell-surface receptors called pattern recognition receptors (PRRs), as well as their known downstream signalling components. Using 350 genome sequences spanning red algae, green algae, and land plant lineages, they compared the two classes of surface-receptors: receptor-like kinases (RLKs) and receptor-like proteins (RLPs). The group found that RLPs emerged earlier than RLKs, and that the LRR-RLKs and LRR-RLPs showed notable expansions throughout different plant lineages – likely due to their involvement in pathogen detection. Focusing on the largest PRR family, the LRR-RLPs, the authors found a key conserved domain architecture: an island domain (ID) + 4 LRR motifs. Interestingly, they also noticed that the ID + 4 LRRs is also present in the LRR-RLK subgroup Xb, which includes several receptors involved in developmental processes. By performing several domain swapping experiments, they identified regions in the LRR-RLP and LRR-RLK-Xb families responsible for both immune and development pathway activation. The authors propose a persuasive model of “modular evolution,” where specific domains of cell-surface receptors evolve to recognize diverse ligands while other domains remain conserved to preserve distinct downstream signalling pathways. (Summary by Tamar Av-Shalom) Nat Commun. 10.1038/s41467-023-44408-3
Seasonal flowering and seasonal growth measure light duration differently
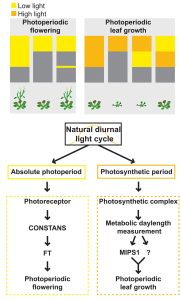 One of the first lessons a plant biologist learns is that many plants coordinate their seasonal flowering through measuring daylength, and that this process involves both photoreceptors and the biological clock that functions inside of cells. Of course, daylength also affects plant metabolism, in part by changing the amount of light received for photosynthesis, but also through integration with the clock. Here, Wang et al. carried out an elegant set of experiments to investigate the interactions between light, photoperiod, and seasonal growth. They identified MIPS-1 (MYOINOSITOL-1-PHOSPHATE SYNTHASE 1), a gene encoding an enzyme that produces myo-inositol, as photoperiod sensitive and required for growth in long but not short days; in other words, as a marker for seasonal growth. Analysis of mips1 mutants revealed that the growth response to photoperiod is genetically distinct from the flowering response to photoperiod. Interestingly, the daylength control of flowering responds to very low light levels, whereas the seasonal growth response is less sensitive to low light levels, and not so much daylength sensitive as “photosynthesis duration” sensitive. These results reveal that plants use multiple systems to measure different aspects of daylength and season and modulate their responses accordingly. It will be very interesting to investigate these new findings in crop plants. (Summary by Mary Williams @PlantTeaching) 10.1126/science.adg9196
One of the first lessons a plant biologist learns is that many plants coordinate their seasonal flowering through measuring daylength, and that this process involves both photoreceptors and the biological clock that functions inside of cells. Of course, daylength also affects plant metabolism, in part by changing the amount of light received for photosynthesis, but also through integration with the clock. Here, Wang et al. carried out an elegant set of experiments to investigate the interactions between light, photoperiod, and seasonal growth. They identified MIPS-1 (MYOINOSITOL-1-PHOSPHATE SYNTHASE 1), a gene encoding an enzyme that produces myo-inositol, as photoperiod sensitive and required for growth in long but not short days; in other words, as a marker for seasonal growth. Analysis of mips1 mutants revealed that the growth response to photoperiod is genetically distinct from the flowering response to photoperiod. Interestingly, the daylength control of flowering responds to very low light levels, whereas the seasonal growth response is less sensitive to low light levels, and not so much daylength sensitive as “photosynthesis duration” sensitive. These results reveal that plants use multiple systems to measure different aspects of daylength and season and modulate their responses accordingly. It will be very interesting to investigate these new findings in crop plants. (Summary by Mary Williams @PlantTeaching) 10.1126/science.adg9196
Chemical defense: Exploring two-component plant defense mechanisms in Panax species
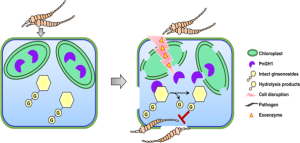 In the interesting world of plant defenses, plants have secret weapons called defense metabolites that stay quiet until a pathogen comes knocking. Plants have evolved two-component chemical defence systems to protect against pathogens while striking a balance between growth promotion and defence mechanisms. Two-component chemical defense systems are coined as such due to the separation of the defensive enzyme from its substrate. Upon activation, these components mix to form the protective molecule; cyanogenic glycosides are a well-known example of this concept. Here, Ma et al. revealed a two-component chemical defense system in Panax notoginseng against the necrotrophic fungus Mycocentrospora acerina. Ginsenosides, widespread secondary metabolites in Panax sp., are synthesized using substantial resources by P. notoginseng, hinting at vital plant functions or benefits. This plant defense mechanism is comprised of PnGH1, a chloroplast-localized β-glucosidase, and its partnering substrates, the 20(S)-protopanaxadiol (PPD) type ginsenosides. Upon cell disruption, induced exoenzymes from pathogenic fungi facilitate the release and blending of chloroplast-localized PnGH1 with its substrates, resulting in heightened potency of the hydrolysis products in inhibiting pathogen growth, both in vitro and in vivo, compared to non-hydrolyzed substrates. This defense mechanism extends beyond P. notoginseng, encompassing Panax quinquefolium and Panax ginseng. Understanding the mechanisms involved in plant defense allows researchers to design pesticides that can specifically target and disrupt the pathways utilized by pathogens. (Summary by Tuyelee Das; @das_tuyelee) Nature Comms. 10.1038/s41467-024-44854-7
In the interesting world of plant defenses, plants have secret weapons called defense metabolites that stay quiet until a pathogen comes knocking. Plants have evolved two-component chemical defence systems to protect against pathogens while striking a balance between growth promotion and defence mechanisms. Two-component chemical defense systems are coined as such due to the separation of the defensive enzyme from its substrate. Upon activation, these components mix to form the protective molecule; cyanogenic glycosides are a well-known example of this concept. Here, Ma et al. revealed a two-component chemical defense system in Panax notoginseng against the necrotrophic fungus Mycocentrospora acerina. Ginsenosides, widespread secondary metabolites in Panax sp., are synthesized using substantial resources by P. notoginseng, hinting at vital plant functions or benefits. This plant defense mechanism is comprised of PnGH1, a chloroplast-localized β-glucosidase, and its partnering substrates, the 20(S)-protopanaxadiol (PPD) type ginsenosides. Upon cell disruption, induced exoenzymes from pathogenic fungi facilitate the release and blending of chloroplast-localized PnGH1 with its substrates, resulting in heightened potency of the hydrolysis products in inhibiting pathogen growth, both in vitro and in vivo, compared to non-hydrolyzed substrates. This defense mechanism extends beyond P. notoginseng, encompassing Panax quinquefolium and Panax ginseng. Understanding the mechanisms involved in plant defense allows researchers to design pesticides that can specifically target and disrupt the pathways utilized by pathogens. (Summary by Tuyelee Das; @das_tuyelee) Nature Comms. 10.1038/s41467-024-44854-7
An opportunistic plant pathogen disrupts leaf microbiome through secretion of plant cell wall degrading enzymes
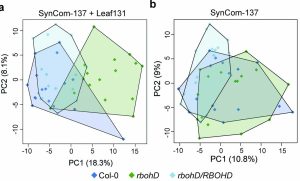 Healthy plants possess a functional immune system, and their leaves harbor a structured microbial community, including opportunistic pathogens. These opportunistic pathogens can trigger plant diseases under conducive conditions, such as when plant immunity is compromised, and the microbial community is disrupted. It remains elusive whether and how an opportunistic pathogen can directly disrupt the microbiome to cause disease. By using an immunocompromised Arabidopsis genotype, its complementation line, and a leaf-derived bacterial synthetic community (SynCom), Pfeilmeier et al. established a direct causative role of an opportunistic pathogen (Xanthomonas Leaf 131) in perturbing leaf microbial communities for disease development. By adopting a quantitative assay in Arabidopsis leaves, the authors confirmed leaf tissue degradation as a potential mode of virulence for this pathogenic strain. Further, they generated targeted deletion and mutated Xanthomonas Leaf 131 strains to provide both in vitro and in planta evidence that T2SS (type II secretions systems) secreted plant cell-wall degrading enzymes cause this characteristic leaf tissue degradation and favor specific commensal bacteria to perturb the leaf microbiome. This study revealed a mechanism of context dependent transition from commensalism to pathogenic behavior of an opportunistic pathogen by direct perturbation of the leaf microbiome. (Summary by Arijit Mukherjee ArijitM61745830) Nat. Microbiol. 10.1038/s41564-023-01555-z
Healthy plants possess a functional immune system, and their leaves harbor a structured microbial community, including opportunistic pathogens. These opportunistic pathogens can trigger plant diseases under conducive conditions, such as when plant immunity is compromised, and the microbial community is disrupted. It remains elusive whether and how an opportunistic pathogen can directly disrupt the microbiome to cause disease. By using an immunocompromised Arabidopsis genotype, its complementation line, and a leaf-derived bacterial synthetic community (SynCom), Pfeilmeier et al. established a direct causative role of an opportunistic pathogen (Xanthomonas Leaf 131) in perturbing leaf microbial communities for disease development. By adopting a quantitative assay in Arabidopsis leaves, the authors confirmed leaf tissue degradation as a potential mode of virulence for this pathogenic strain. Further, they generated targeted deletion and mutated Xanthomonas Leaf 131 strains to provide both in vitro and in planta evidence that T2SS (type II secretions systems) secreted plant cell-wall degrading enzymes cause this characteristic leaf tissue degradation and favor specific commensal bacteria to perturb the leaf microbiome. This study revealed a mechanism of context dependent transition from commensalism to pathogenic behavior of an opportunistic pathogen by direct perturbation of the leaf microbiome. (Summary by Arijit Mukherjee ArijitM61745830) Nat. Microbiol. 10.1038/s41564-023-01555-z