Plant Science Research Weekly: February 10, 2023
Review: Hypes, hopes, and the way forward for microalgal biotechnology
 Microalgae are small, photosynthetic algae that have emerged as important contributors to food and nutrient production. This review describes the many ways they can be grown (e.g., autotrophic, with sunlight or artificial light; heterotrophic, with sugar inputs; or mixotrophic, a combination of both) and the many things they can produce. Interestingly, the mixotrophic method can be quite efficient in part because the O2 evolved during photosynthesis is captured for use in respiration and the CO2 is similarly recycled, decreasing the need to de-gas the cultures. Per unit of area, microalgae can greatly outproduce crop plants, and can be sources of protein, carbohydrates and/or lipids (including “fish” oils). They are also amenable to genetic engineering so can produce high-value products such as oral vaccines or antibodies. The review provides calculations for efficiencies considering various inputs and outputs, as well as the advantages and disadvantages compared to other systems for the production of protein therapeutics, demonstrating the current potential of microalgal methods. Further, the authors observe, “There is room for development”. (Summary by Mary Williams @PlantTeaching) Trends Biotechnol.10.1016/j.tibtech.2022.12.017
Microalgae are small, photosynthetic algae that have emerged as important contributors to food and nutrient production. This review describes the many ways they can be grown (e.g., autotrophic, with sunlight or artificial light; heterotrophic, with sugar inputs; or mixotrophic, a combination of both) and the many things they can produce. Interestingly, the mixotrophic method can be quite efficient in part because the O2 evolved during photosynthesis is captured for use in respiration and the CO2 is similarly recycled, decreasing the need to de-gas the cultures. Per unit of area, microalgae can greatly outproduce crop plants, and can be sources of protein, carbohydrates and/or lipids (including “fish” oils). They are also amenable to genetic engineering so can produce high-value products such as oral vaccines or antibodies. The review provides calculations for efficiencies considering various inputs and outputs, as well as the advantages and disadvantages compared to other systems for the production of protein therapeutics, demonstrating the current potential of microalgal methods. Further, the authors observe, “There is room for development”. (Summary by Mary Williams @PlantTeaching) Trends Biotechnol.10.1016/j.tibtech.2022.12.017
Opinion – From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome
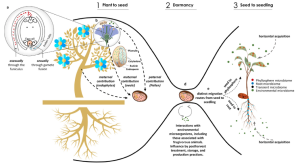 Seeds not only carry the embryo that will germinate and give rise to a new individual, but they are also the vector to the next generation for microorganisms from the mother plant and the environment. Consequently, seeds link the microbiome between plant generations. In this thorough review, Abdelfattah and colleagues take us on the journey of microbial inheritance in seed plants, which starts from the very beginning of seed development. Seeds interact with their surroundings, receiving microorganisms from the air, the rain and –potentially– from the ovule and sperm cells. When seeds are dispersed and reach the soil, they are exposed to entirely new microbiota, from the soil and possibly from the dispersers’ guts. When germination occurs, seed microbial communities will migrate to their final destination, either in the roots or the shoot, each with a distinctive composition. Some questions about the processes and mechanisms involved in this journey are still a matter of debate. However, this paper provides a fascinating overview of the processes that shape microbial inheritance through seeds, making it obligate reading for those interested in the subject. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Trends Microbiol. 10.1016/j.tim.2022.10.009
Seeds not only carry the embryo that will germinate and give rise to a new individual, but they are also the vector to the next generation for microorganisms from the mother plant and the environment. Consequently, seeds link the microbiome between plant generations. In this thorough review, Abdelfattah and colleagues take us on the journey of microbial inheritance in seed plants, which starts from the very beginning of seed development. Seeds interact with their surroundings, receiving microorganisms from the air, the rain and –potentially– from the ovule and sperm cells. When seeds are dispersed and reach the soil, they are exposed to entirely new microbiota, from the soil and possibly from the dispersers’ guts. When germination occurs, seed microbial communities will migrate to their final destination, either in the roots or the shoot, each with a distinctive composition. Some questions about the processes and mechanisms involved in this journey are still a matter of debate. However, this paper provides a fascinating overview of the processes that shape microbial inheritance through seeds, making it obligate reading for those interested in the subject. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Trends Microbiol. 10.1016/j.tim.2022.10.009
Structural and functional insights into the chloroplast division site regulators PARC6 and PDV1 in the intermembrane space
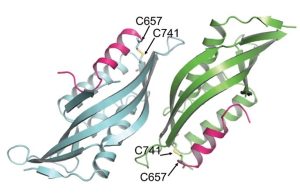 Chloroplasts divide by binary fission driven by a protein ring, the position of which is regulated by the Min system (derived from the system in bacteria). The inner envelope membrane protein PARC6 (PARALOG OF ARC6) is a key component. Here Sun et al. generated crystal structures showing that PARC6 interacts with the outer envelope membrane protein PDV1 (PLASTID DIVISION1). There are two interaction sites, one inside a pocket-like structure of PARC6 and one in a hydrophobic region on the lid of PARC6. Interaction at both sites leads to PARC6 dimerization, which is needed for correct protein function and chloroplast division. This PARC6-PDV1 interaction is redox regulated. Under oxidizing conditions, a disulfide bond within PARC6 blocks the binding pocket and inhibits the PDV1 interaction. In vitro, the reducing agent DTT reduces this bond to permit the PARC6-PDV1 interaction. In planta, this same reaction is triggered upon transition to light, driven by the accumulation of reductants. Therefore, this dynamic PARC6-PDV1 interaction ensures that chloroplast division is coordinated with light. (Summary by Rose McNelly @RoseMcN) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2215575120
Chloroplasts divide by binary fission driven by a protein ring, the position of which is regulated by the Min system (derived from the system in bacteria). The inner envelope membrane protein PARC6 (PARALOG OF ARC6) is a key component. Here Sun et al. generated crystal structures showing that PARC6 interacts with the outer envelope membrane protein PDV1 (PLASTID DIVISION1). There are two interaction sites, one inside a pocket-like structure of PARC6 and one in a hydrophobic region on the lid of PARC6. Interaction at both sites leads to PARC6 dimerization, which is needed for correct protein function and chloroplast division. This PARC6-PDV1 interaction is redox regulated. Under oxidizing conditions, a disulfide bond within PARC6 blocks the binding pocket and inhibits the PDV1 interaction. In vitro, the reducing agent DTT reduces this bond to permit the PARC6-PDV1 interaction. In planta, this same reaction is triggered upon transition to light, driven by the accumulation of reductants. Therefore, this dynamic PARC6-PDV1 interaction ensures that chloroplast division is coordinated with light. (Summary by Rose McNelly @RoseMcN) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2215575120
Green means go: Green light promotes hypocotyl elongation via brassinosteroid signaling
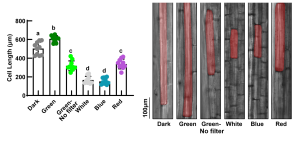 Generally, light inhibits hypocotyl elongation. Like red and blue light, green light was previously reported to inhibit hypocotyl elongation in several plants. Here, Hao et al. discovered that the inhibition of hypocotyl growth by green light is due to wavelength impurities in the green lights used. Commercial green LED lights of 525 nm and 550 nm wavelengths generate blue and red light tails respectively, which constitute impurities, hence inhibit hypocotyl growth. Pure green light of 550 nm (Green 550) promotes hypocotyl elongation. However, like other light wavelengths, it also promotes apical hook opening and cotyledon separation. The influence of Green 550 was shown not to be photosynthesis dependent or due to effects on germination. Green 500 promotes hypocotyl elongation via the brassinosteroid signaling pathway and acts as shade signals for plants to adapt under canopy. The authors stipulated that Green 500 effects on hypocotyl elongation may not be mediated by known photoreceptor, suggesting a need to identify a green light receptor. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Cell 10.1093/plcell/koad022
Generally, light inhibits hypocotyl elongation. Like red and blue light, green light was previously reported to inhibit hypocotyl elongation in several plants. Here, Hao et al. discovered that the inhibition of hypocotyl growth by green light is due to wavelength impurities in the green lights used. Commercial green LED lights of 525 nm and 550 nm wavelengths generate blue and red light tails respectively, which constitute impurities, hence inhibit hypocotyl growth. Pure green light of 550 nm (Green 550) promotes hypocotyl elongation. However, like other light wavelengths, it also promotes apical hook opening and cotyledon separation. The influence of Green 550 was shown not to be photosynthesis dependent or due to effects on germination. Green 500 promotes hypocotyl elongation via the brassinosteroid signaling pathway and acts as shade signals for plants to adapt under canopy. The authors stipulated that Green 500 effects on hypocotyl elongation may not be mediated by known photoreceptor, suggesting a need to identify a green light receptor. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Cell 10.1093/plcell/koad022
Underwater blues: Molecular rewiring of stomatal development in Rorippa aquatica
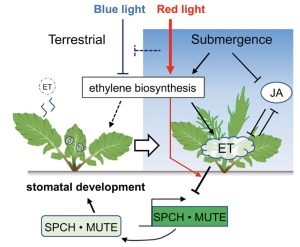 Unanticipated flooding conditions challenge the survival and overall growth and development of a plant, for example stomatal development is suppressed under submerged conditions. Stomata are microscopic pores on the surface of leaves of a plant that play an important role in the exchange of gases between the plant and the atmosphere. Ikematsu et al. outline a molecular framework underlying the inhibition of stomatal development under water by using an amphibious plant Rorippa aquatica, an emerging model in the Brassicaceae family. Rorippa aquatica produces different leaf forms (heterophylly) when submerged as opposed to in dry air, with the submerged leaves also showing fewer stomata. Their findings suggest that submergence-triggered inhibition of stomata can be uncoupled from morphological heterophylly. Upon submergence there is reprogramming of a suite of genes underlying light signaling and hormones, eventually repressing the stomatal developmental genes SPCH and MUTE. The authors found that blue light promotes, whereas red light inhibits, stomatal development. Their results indicate that in these plants there has been a rewiring of the pathway from red light to ethylene. (Summary by Arpita Yadav @arpita_yadav_). Curr. Biol. 10.1016/j.cub.2022.12.064
Unanticipated flooding conditions challenge the survival and overall growth and development of a plant, for example stomatal development is suppressed under submerged conditions. Stomata are microscopic pores on the surface of leaves of a plant that play an important role in the exchange of gases between the plant and the atmosphere. Ikematsu et al. outline a molecular framework underlying the inhibition of stomatal development under water by using an amphibious plant Rorippa aquatica, an emerging model in the Brassicaceae family. Rorippa aquatica produces different leaf forms (heterophylly) when submerged as opposed to in dry air, with the submerged leaves also showing fewer stomata. Their findings suggest that submergence-triggered inhibition of stomata can be uncoupled from morphological heterophylly. Upon submergence there is reprogramming of a suite of genes underlying light signaling and hormones, eventually repressing the stomatal developmental genes SPCH and MUTE. The authors found that blue light promotes, whereas red light inhibits, stomatal development. Their results indicate that in these plants there has been a rewiring of the pathway from red light to ethylene. (Summary by Arpita Yadav @arpita_yadav_). Curr. Biol. 10.1016/j.cub.2022.12.064
BIN2 signaling network constructed using proximity labeling and phosphoproteomics methods
 The brassinosteroid (BR) signaling pathway is essential for plant growth and development. BRASSINOSTEROID-INSENSITIVE2 (BIN2) is a GSK3-like kinase and a repressor of the BR signaling pathway. In the absence of BR, BIN2 phosphorylates BRASSINAZOLE-RESISTANT1 (BZR1) family transcription factors to inactivate them and retain them in the cytoplasm. Upon the perception of BR by its receptor kinase BRI1, BIN2 is deactivated through dephosphorylation, ubiquitination and degradation, which releases the inactivation of BZR1. Beyond that, BIN2 has been recently reported to have broader roles in the crosstalk with other hormone signaling pathways and stress responses. Thus, it is important to identify the substrates of BIN2. However, the interaction between kinases and substrates is hard to capture, due to their transient and dynamic nature. In this recent publication, Kim et al. present huge advances in mapping the BIN2 signaling network. Using TurboID-based proximity labeling (TbPL) followed by mass spectrometry, 482 proteins were identified to be proximal to BIN2. Using BIN2 inhibitor treatment and phosphoproteomics, over one-third of these proteins were found to be dephosphorylated upon inhibition of BIN2 and considered as BIN2 substrates. These interactors and substrates of BIN2 were constructed into a BIN2 signaling network. (Summary by Xiaohui Li @Xiao_hui_Li) Plant Cell 10.1093/plcell/koad013
The brassinosteroid (BR) signaling pathway is essential for plant growth and development. BRASSINOSTEROID-INSENSITIVE2 (BIN2) is a GSK3-like kinase and a repressor of the BR signaling pathway. In the absence of BR, BIN2 phosphorylates BRASSINAZOLE-RESISTANT1 (BZR1) family transcription factors to inactivate them and retain them in the cytoplasm. Upon the perception of BR by its receptor kinase BRI1, BIN2 is deactivated through dephosphorylation, ubiquitination and degradation, which releases the inactivation of BZR1. Beyond that, BIN2 has been recently reported to have broader roles in the crosstalk with other hormone signaling pathways and stress responses. Thus, it is important to identify the substrates of BIN2. However, the interaction between kinases and substrates is hard to capture, due to their transient and dynamic nature. In this recent publication, Kim et al. present huge advances in mapping the BIN2 signaling network. Using TurboID-based proximity labeling (TbPL) followed by mass spectrometry, 482 proteins were identified to be proximal to BIN2. Using BIN2 inhibitor treatment and phosphoproteomics, over one-third of these proteins were found to be dephosphorylated upon inhibition of BIN2 and considered as BIN2 substrates. These interactors and substrates of BIN2 were constructed into a BIN2 signaling network. (Summary by Xiaohui Li @Xiao_hui_Li) Plant Cell 10.1093/plcell/koad013
Tackling a long-standing question about floral development
 Floral development has been the object of decades of plant research, yet many fundamental questions remain. One of these is the mechanism by which LEAFY (LFY), the master transcription factor of floral development, works with UNUSUAL FLORAL ORGANS (UFO), an F-box protein, to regulate petal and stamen development. In Arabidopsis (Arabidopsis thaliana), LFY and UFO work together to regulate APETALA3 (AP3), and UFO has been thought to be involved in the degradation of LFY. A recent paper by Rieu et al. tackled this question and showed that UFO works as a transcriptional cofactor of LFY to regulate AP3 expression, and that its F-box domain is dispensable for the transcriptional activity of the LFY-UFO complex. UFO allows LFY to bind to new genomic sites that the transcription factor cannot bind on its own, and the cryogenic electron microscopy (cryo-EM) structure of the (ASK1)-UFO-LFY-DNA complex revealed that UFO directly interacts with DNA. If you are wondering whether this mechanism is only found in Arabidopsis, the authors show that several LFY proteins from different species can interact (and induce gene expression) with Arabidopsis UFO. These results suggest that the LFY-UFO interaction may be conserved beyond flowering plants. Interacting with non-transcription factor proteins could be a more widespread mechanism than is currently thought to fine-tune gene regulation and modulate transcription factor activity in different territories. (Summary by Laura Turchi @turchi_l) Nature Plants 10.1038/s41477-022-01336-2
Floral development has been the object of decades of plant research, yet many fundamental questions remain. One of these is the mechanism by which LEAFY (LFY), the master transcription factor of floral development, works with UNUSUAL FLORAL ORGANS (UFO), an F-box protein, to regulate petal and stamen development. In Arabidopsis (Arabidopsis thaliana), LFY and UFO work together to regulate APETALA3 (AP3), and UFO has been thought to be involved in the degradation of LFY. A recent paper by Rieu et al. tackled this question and showed that UFO works as a transcriptional cofactor of LFY to regulate AP3 expression, and that its F-box domain is dispensable for the transcriptional activity of the LFY-UFO complex. UFO allows LFY to bind to new genomic sites that the transcription factor cannot bind on its own, and the cryogenic electron microscopy (cryo-EM) structure of the (ASK1)-UFO-LFY-DNA complex revealed that UFO directly interacts with DNA. If you are wondering whether this mechanism is only found in Arabidopsis, the authors show that several LFY proteins from different species can interact (and induce gene expression) with Arabidopsis UFO. These results suggest that the LFY-UFO interaction may be conserved beyond flowering plants. Interacting with non-transcription factor proteins could be a more widespread mechanism than is currently thought to fine-tune gene regulation and modulate transcription factor activity in different territories. (Summary by Laura Turchi @turchi_l) Nature Plants 10.1038/s41477-022-01336-2
MicroRNA408 and its encoded peptide regulate sulfur assimilation and arsenic stress response in Arabidopsis
 MicroRNAs (miRNAs) are small non-coding RNAs that down-regulate their targets through translational repression or mRNA cleavage. Some pri-miRNAs encode small regulatory peptides (miPEP) which regulate plant growth and development by modulating subsequent miRNA expression. How different biotic or abiotic stresses may affect the role of these peptides remains unclear. In this work, Kumar et al. focus on the role of miR408 and its encoded peptide (miPEP408) in response to heavy metal and nutrient deficiency. The authors developed CRISPR/Cas9 and overexpressing lines to unravel its role. Under sulfate deficit and As stress, the overexpressing lines showed a more sensitive phenotype, while CRISPR/Cas9 edited lines exhibited increased root length and fresh weight. Application of miPEP408 recovered the WT phenotype in CRISPR/Cas9 edited lines. The authors propose a model in which under sulfate deficit and As stress, miPEP408 regulates miR408 which downregulates glutathione S-transferase (GSTU25) and sulfur reduction genes in Arabidopsis. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiad033
MicroRNAs (miRNAs) are small non-coding RNAs that down-regulate their targets through translational repression or mRNA cleavage. Some pri-miRNAs encode small regulatory peptides (miPEP) which regulate plant growth and development by modulating subsequent miRNA expression. How different biotic or abiotic stresses may affect the role of these peptides remains unclear. In this work, Kumar et al. focus on the role of miR408 and its encoded peptide (miPEP408) in response to heavy metal and nutrient deficiency. The authors developed CRISPR/Cas9 and overexpressing lines to unravel its role. Under sulfate deficit and As stress, the overexpressing lines showed a more sensitive phenotype, while CRISPR/Cas9 edited lines exhibited increased root length and fresh weight. Application of miPEP408 recovered the WT phenotype in CRISPR/Cas9 edited lines. The authors propose a model in which under sulfate deficit and As stress, miPEP408 regulates miR408 which downregulates glutathione S-transferase (GSTU25) and sulfur reduction genes in Arabidopsis. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiad033
Bitter gourd protein for potential defense against plant viral diseases
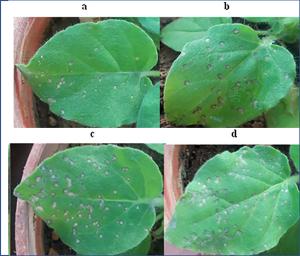 Bitter gourds (or bitter melon) have been a childhood nightmare of mine. I remember that as a child, my grandparents used to force me to consume these really bitter gourds (Momordica charantia) telling me that they had ‘many benefits’. In addition to being a rich source of antioxidants, M. charantia seeds and fruit contain a ribosome inactivating protein (RIPs) called MAP30 (Momordica Antiviral Protein 30 kDa), known for its anti-tumor and anti-HIV activity as well as anti-microbial activity. RIPs chemically affect the universally conserved α-sarcin loop of large rRNAs, thereby inactivating the ribosome and translation. In a new study, Amirzadeh et al. showed that recombinant MAP30 (rMAP30) can suppress tobacco mosaic virus (TMV) and cucumber mosaic virus (CMW) infection in Nicotiana glutinousa and Chenopodium quinoa seedlings. rMAP30 significantly reduced the number of infection spots on leaves by 60-70% These in vivo anti-viral properties showed potential applications of rMAP30 in the field to control bacterial and virus infections and consequently crop losses. However, more in vivo studies with other viruses and with mature plants are needed to confirm these results. In reality, it might not be feasible to inoculate every crop leaf in the field. Hence, further studies need to be done to see whether different inoculation techniques such as aerosolized rMAP30 or rMAP30 in irrigation water can reduce crop diseases. (Summary by Apple Chew @_applechew) Sci. Rep. 10.1038/s41598-023-29365-7
Bitter gourds (or bitter melon) have been a childhood nightmare of mine. I remember that as a child, my grandparents used to force me to consume these really bitter gourds (Momordica charantia) telling me that they had ‘many benefits’. In addition to being a rich source of antioxidants, M. charantia seeds and fruit contain a ribosome inactivating protein (RIPs) called MAP30 (Momordica Antiviral Protein 30 kDa), known for its anti-tumor and anti-HIV activity as well as anti-microbial activity. RIPs chemically affect the universally conserved α-sarcin loop of large rRNAs, thereby inactivating the ribosome and translation. In a new study, Amirzadeh et al. showed that recombinant MAP30 (rMAP30) can suppress tobacco mosaic virus (TMV) and cucumber mosaic virus (CMW) infection in Nicotiana glutinousa and Chenopodium quinoa seedlings. rMAP30 significantly reduced the number of infection spots on leaves by 60-70% These in vivo anti-viral properties showed potential applications of rMAP30 in the field to control bacterial and virus infections and consequently crop losses. However, more in vivo studies with other viruses and with mature plants are needed to confirm these results. In reality, it might not be feasible to inoculate every crop leaf in the field. Hence, further studies need to be done to see whether different inoculation techniques such as aerosolized rMAP30 or rMAP30 in irrigation water can reduce crop diseases. (Summary by Apple Chew @_applechew) Sci. Rep. 10.1038/s41598-023-29365-7
Does fire drive fatty acid composition in seed coats of physically dormant species?
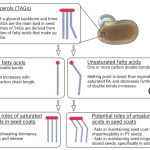
 Physical dormancy is imposed by an impermeable seed coat that prevents water uptake. For germination to occur, this coat must be broken. Fire passage, for example, breaks physical dormancy in ecosystems where such events are recurrent. However, the mechanisms behind how fire achieves this has yet to be explored. Fatty acids (FAs) influence seed coat permeability, so they are a promising starting point for elucidating the chemical mechanism behind physical dormancy break. In this pioneering paper, McInnes and colleagues assess the FAs composition of seeds from 25 Fabaceae from fire-prone and fire-free Australian ecosystems. Species from fire-prone ecosystems had seeds with relatively more FAs than those from fire-free sites. Notably, these species had larger FAs and relatively more unsaturated ones. Large, unsaturated FAs increase seed coat impermeability and have higher melting points that match the temperatures required for the germination of species from fire-prone ecosystems. This research ignites a new, fascinating research area that will contribute to a better understanding of the germination of physically dormant seeds. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Biol. 10.1111/plb.13498
Physical dormancy is imposed by an impermeable seed coat that prevents water uptake. For germination to occur, this coat must be broken. Fire passage, for example, breaks physical dormancy in ecosystems where such events are recurrent. However, the mechanisms behind how fire achieves this has yet to be explored. Fatty acids (FAs) influence seed coat permeability, so they are a promising starting point for elucidating the chemical mechanism behind physical dormancy break. In this pioneering paper, McInnes and colleagues assess the FAs composition of seeds from 25 Fabaceae from fire-prone and fire-free Australian ecosystems. Species from fire-prone ecosystems had seeds with relatively more FAs than those from fire-free sites. Notably, these species had larger FAs and relatively more unsaturated ones. Large, unsaturated FAs increase seed coat impermeability and have higher melting points that match the temperatures required for the germination of species from fire-prone ecosystems. This research ignites a new, fascinating research area that will contribute to a better understanding of the germination of physically dormant seeds. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant Biol. 10.1111/plb.13498