Plant Science Research Weekly: Feb. 19, 2021
Review: Time to wake up: Epigenetic and small-RNA-mediated regulation during seed germination
 Seed germination is a crucial step in plants’ life cycle and has critical implications in ecological and agronomic contexts. Therefore, there has been an increasing interest in understanding the mechanisms that prevent or trigger this process. Here, Luján-Soto and Dinkova provide a thorough review of the molecular mechanisms that regulate germination, focusing on histone modifications, DNA methylation, and small non-coding RNAs (sRNAs). An overall reduced transcriptional activity characterizes the final stages of seed development. Still, the expression of maturation and dormancy-related genes (e.g., LEAFY COTYLEDON 1 and 2 and DELAY OF GERMINATION-1) increases during the same stages thanks to the effects of histone modifications like H3K4me3. When imbibition starts, these maturation/dormancy genes are silenced by the action of H3K27me3, and the transcriptional landscape is modified to stimulate germination and seedling growth. To achieve this, the DNA experiences a general demethylation that results in reduced sensitivity to ABA, the promoter of seed dormancy. Recent research has shown that different sRNAs have a key role in shaping seed genetic programs. Still, their role in this process has seldom been explored. Given the detailed overview it provides, this review is an obligate reading for everyone interested in the molecular basis behind seed germination regulation. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plants 10.3390/plants10020236
Seed germination is a crucial step in plants’ life cycle and has critical implications in ecological and agronomic contexts. Therefore, there has been an increasing interest in understanding the mechanisms that prevent or trigger this process. Here, Luján-Soto and Dinkova provide a thorough review of the molecular mechanisms that regulate germination, focusing on histone modifications, DNA methylation, and small non-coding RNAs (sRNAs). An overall reduced transcriptional activity characterizes the final stages of seed development. Still, the expression of maturation and dormancy-related genes (e.g., LEAFY COTYLEDON 1 and 2 and DELAY OF GERMINATION-1) increases during the same stages thanks to the effects of histone modifications like H3K4me3. When imbibition starts, these maturation/dormancy genes are silenced by the action of H3K27me3, and the transcriptional landscape is modified to stimulate germination and seedling growth. To achieve this, the DNA experiences a general demethylation that results in reduced sensitivity to ABA, the promoter of seed dormancy. Recent research has shown that different sRNAs have a key role in shaping seed genetic programs. Still, their role in this process has seldom been explored. Given the detailed overview it provides, this review is an obligate reading for everyone interested in the molecular basis behind seed germination regulation. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plants 10.3390/plants10020236
A novel hyperactive UVR8 variant for enhanced UV-B responses
 Plants perceive UV-B radiation using a specialized photoreceptor protein called UVR8. Upon UV-B exposure, activated UVR8 initiates a signaling cascade, inducing a range of morphological and biochemical responses that facilitates UV adaptation. In a mutagenesis screen, Podolec et al. identified a mutant named uvr8-17D, which produces a hyperactive variant of UVR8, harbouring a glycine-101 to serine change (G101S). The mutant seedlings, although grew normally under white light, exhibited hypersensitive phenotypes and enhanced survivability under UV-B radiation. Generally, UVR8 proteins are inactive homodimers in their ground state and become active monomers when exposed to UV-B. Interestingly, UVR8G101S encoded by uvr8-17D is constitutively monomeric, making it weakly active even in the absence of UV-B. Under UV-B, UVR8G101S is imported more into the nucleus due to its enhanced interaction with COP1. UVR8 photocycle becomes complete only when the monomers are recruited and brought back into inactive homodimers by RUP1 and RUP2. In uvr8-17D, this photocycle remains incomplete due to the inability of UVR8G101S to form homodimers. Detailed structural analysis revealed that the G101S mutation in UVR8 distorts the conformation of a loop that is crucial for its homodimerization. Together, the enhanced UV-B signaling by this novel hyperactive UVR8 variant increases the sensitivity and response of plants to UV-B radiation, which can be highly useful for fundamental research as well as crop improvement programs in future. (Summary by Yadukrishnan Premachandran @yadukrishprem) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2017284118
Plants perceive UV-B radiation using a specialized photoreceptor protein called UVR8. Upon UV-B exposure, activated UVR8 initiates a signaling cascade, inducing a range of morphological and biochemical responses that facilitates UV adaptation. In a mutagenesis screen, Podolec et al. identified a mutant named uvr8-17D, which produces a hyperactive variant of UVR8, harbouring a glycine-101 to serine change (G101S). The mutant seedlings, although grew normally under white light, exhibited hypersensitive phenotypes and enhanced survivability under UV-B radiation. Generally, UVR8 proteins are inactive homodimers in their ground state and become active monomers when exposed to UV-B. Interestingly, UVR8G101S encoded by uvr8-17D is constitutively monomeric, making it weakly active even in the absence of UV-B. Under UV-B, UVR8G101S is imported more into the nucleus due to its enhanced interaction with COP1. UVR8 photocycle becomes complete only when the monomers are recruited and brought back into inactive homodimers by RUP1 and RUP2. In uvr8-17D, this photocycle remains incomplete due to the inability of UVR8G101S to form homodimers. Detailed structural analysis revealed that the G101S mutation in UVR8 distorts the conformation of a loop that is crucial for its homodimerization. Together, the enhanced UV-B signaling by this novel hyperactive UVR8 variant increases the sensitivity and response of plants to UV-B radiation, which can be highly useful for fundamental research as well as crop improvement programs in future. (Summary by Yadukrishnan Premachandran @yadukrishprem) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2017284118
Redox regulation of NADP-malate dehydrogenase is vital under fluctuating light environment
 Redox switches regulate photosynthetic reactions and are mediated by conserved cysteine pairs, activated by reduction under light and deactivated by oxidation in the dark. Those reactions are mediated by thioredoxins (Trx). Usually, only plastidic and not cytosolic isozymes show these redox switches. As an example, the redox-regulated enzyme malate dehydrogenase (MDH) produces malate using NADPH to export reducing products from chloroplasts to the cytosol, which balances the ATP/NADPH ratio in the chloroplast. The activity of MDH in the dark is strictly regulated by two switches at the N and C terminus, suggesting that complete deactivation is necessary to generate acclimation responses. Yokochi et al. used CRISPR-Cas9 to generate mutant plants (MDHΔC-CR) with deleted thiol-based redox switch. The mutants showed no phenotypic changes under standard long-day conditions. However, under short-day or fluctuating light the mutants showed a stunted growth phenotype. The metabolite dynamics in the mutants indicated that the oxidized NADP pool and ferredoxin, which serve as electron acceptors for PSI, are increased in MDHΔC-CR. In conclusion, the thiol-switch redox regulation of MDH was found to be required to maintain plastidic NADPH homeostasis under fluctuating light conditions. (Summary by Camila Ribeiro @camicribeiro86) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2016903118.
Redox switches regulate photosynthetic reactions and are mediated by conserved cysteine pairs, activated by reduction under light and deactivated by oxidation in the dark. Those reactions are mediated by thioredoxins (Trx). Usually, only plastidic and not cytosolic isozymes show these redox switches. As an example, the redox-regulated enzyme malate dehydrogenase (MDH) produces malate using NADPH to export reducing products from chloroplasts to the cytosol, which balances the ATP/NADPH ratio in the chloroplast. The activity of MDH in the dark is strictly regulated by two switches at the N and C terminus, suggesting that complete deactivation is necessary to generate acclimation responses. Yokochi et al. used CRISPR-Cas9 to generate mutant plants (MDHΔC-CR) with deleted thiol-based redox switch. The mutants showed no phenotypic changes under standard long-day conditions. However, under short-day or fluctuating light the mutants showed a stunted growth phenotype. The metabolite dynamics in the mutants indicated that the oxidized NADP pool and ferredoxin, which serve as electron acceptors for PSI, are increased in MDHΔC-CR. In conclusion, the thiol-switch redox regulation of MDH was found to be required to maintain plastidic NADPH homeostasis under fluctuating light conditions. (Summary by Camila Ribeiro @camicribeiro86) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2016903118.
Jasmonate biosynthesis arising from altered cell walls is prompted by turgor-driven mechanical compression
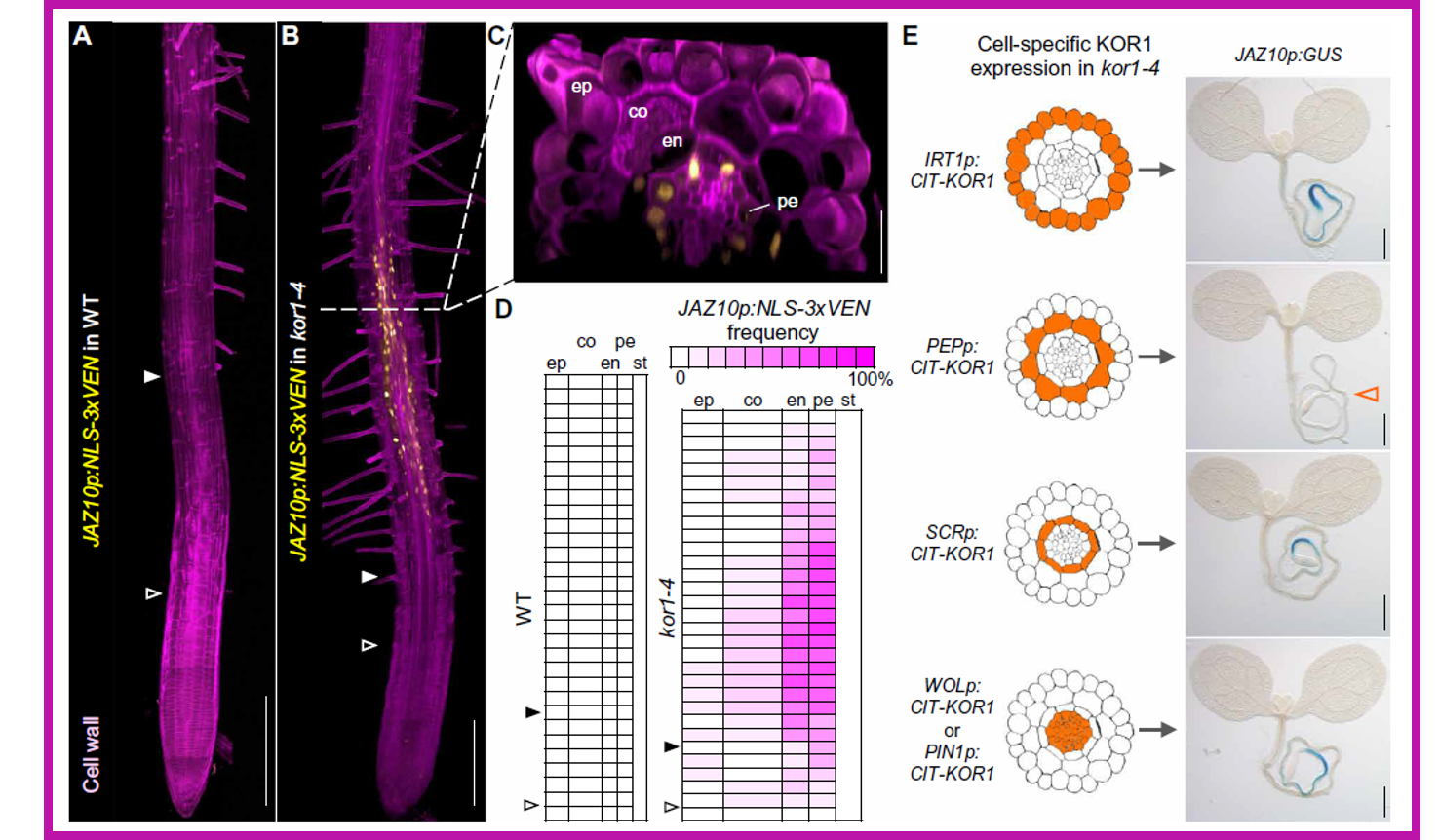 Jasmonate (JA) coordinates plant development and response to a wide range of biotic and abiotic stresses. JA production is stimulated by wounding, which damages cell walls, and increased in plants carrying mutations in cellulose biosynthetic genes. Nevertheless, the intracellular events connecting mechanical stress and JA production largely remain unresolved. In a forward genetic screen aimed at the identification of repressors of JA signaling, Mielke and coworkers isolated mild alleles of korrigan1 (kor1), previously shown to have cellulose deficiencies, that showed activation of JA markers in primary roots and increased JA levels. Besides shorter roots, kor1 mutants displayed enlargement of cortex cells and induction of JA signaling in endodermal and pericycle cells of the early differentiation zone of primary roots. Cortex-specific expression of functional KOR1 complemented the kor1 mutant phenotype and abolished the activation of JA reporters, indicating that KOR1 activity in the cortex prevents JA production in inner tissues of the root. When kor1 mutants were grown on mannitol (hyperosmotic conditions), the mutant phenotype was also reverted to normal, through increased root elongation, reduced size of root cells and abolished JA signaling. These findings suggested that changes in the turgor pressure mediate JA biosynthesis, which is triggered in response to mechanical compression and/or osmotic stress. (Summary by Michela Osnato @michela_osnato) Sci Advances 10.1126/sciadv.abf0356
Jasmonate (JA) coordinates plant development and response to a wide range of biotic and abiotic stresses. JA production is stimulated by wounding, which damages cell walls, and increased in plants carrying mutations in cellulose biosynthetic genes. Nevertheless, the intracellular events connecting mechanical stress and JA production largely remain unresolved. In a forward genetic screen aimed at the identification of repressors of JA signaling, Mielke and coworkers isolated mild alleles of korrigan1 (kor1), previously shown to have cellulose deficiencies, that showed activation of JA markers in primary roots and increased JA levels. Besides shorter roots, kor1 mutants displayed enlargement of cortex cells and induction of JA signaling in endodermal and pericycle cells of the early differentiation zone of primary roots. Cortex-specific expression of functional KOR1 complemented the kor1 mutant phenotype and abolished the activation of JA reporters, indicating that KOR1 activity in the cortex prevents JA production in inner tissues of the root. When kor1 mutants were grown on mannitol (hyperosmotic conditions), the mutant phenotype was also reverted to normal, through increased root elongation, reduced size of root cells and abolished JA signaling. These findings suggested that changes in the turgor pressure mediate JA biosynthesis, which is triggered in response to mechanical compression and/or osmotic stress. (Summary by Michela Osnato @michela_osnato) Sci Advances 10.1126/sciadv.abf0356



