Plant Science Research Weekly: December 4, 2020
Review: Organelles-nucleus-plasmodesmata signaling, and roles in plant physiology, metabolism and stress responses
 Plasmodesmata (PD) are pores that connect plant cells and allow the flow of small molecules and information but also viruses. This traffic is tightly regulated, in large part by the deposition or removal of callose that impedes movement. Azim and Burch-Smith review a key signaling module that controls plasmodesmatal function, that they call the organelle-nucleus-plasmodesmata signaling (OPDS) module. This model stems from genetic studies in which mutants affecting intercellular trafficking were found to have primary defects that affected retrograde (to the nucleus) signaling from mitochondria or plastids. Several other mutants that affect organelle metabolism were also found to affect cell-to-cell movements. Although as yet the signals are largely known, the authors suggest that this signaling module serves to integrate carbon fixation with carbon transport, specifically export to the phloem via PD. They speculate on the evolution of this signaling module. Finally, they address the many questions that remain to understand the OPDS pathway and its contributions to plant physiology and stress responses. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.09.005
Plasmodesmata (PD) are pores that connect plant cells and allow the flow of small molecules and information but also viruses. This traffic is tightly regulated, in large part by the deposition or removal of callose that impedes movement. Azim and Burch-Smith review a key signaling module that controls plasmodesmatal function, that they call the organelle-nucleus-plasmodesmata signaling (OPDS) module. This model stems from genetic studies in which mutants affecting intercellular trafficking were found to have primary defects that affected retrograde (to the nucleus) signaling from mitochondria or plastids. Several other mutants that affect organelle metabolism were also found to affect cell-to-cell movements. Although as yet the signals are largely known, the authors suggest that this signaling module serves to integrate carbon fixation with carbon transport, specifically export to the phloem via PD. They speculate on the evolution of this signaling module. Finally, they address the many questions that remain to understand the OPDS pathway and its contributions to plant physiology and stress responses. (Summary by Mary Williams @PlantTeaching) Curr. Opin. Plant Biol. 10.1016/j.pbi.2020.09.005
Review: Emerging roles of long noncoding RNAs in the cytoplasmic milieu
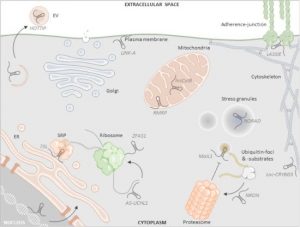 Note: This article focuses on work in mammalian systems but may be of interest to plant biologists. Long non-coding RNA (lncRNAs) are ³200nt RNAs and represent almost 0.2% of cellular RNA pool. Early work focused on their roles in the nucleus, but more recently it has become clear that they also function in the cytoplasm, as reviewed here by Aillaud et al. In the cytoplasm, lncRNAs interact with ribosomes and regulate mRNA translation positively and negatively. They also interact with mitochondria and have roles in cell death and signal transduction during environmental changes, and also regulate endoplasmic reticulum (ER)- dependent autophagy. Various studies have shown the recruitment of lncRNAs with mRNAs in stress granules and with proteins in forming processing bodies. The authors also highlight the involvement of lncRNAs in ubiquitination-dependent protein processing and decay. This review is a great resource on how mammalian lncRNAs affect spatiotemporal responses of cytoplasmic organelles during stress condition. (Summary by Sunita Patak @psunita980) non-coding RNA 10.3390/ncrna6040044
Note: This article focuses on work in mammalian systems but may be of interest to plant biologists. Long non-coding RNA (lncRNAs) are ³200nt RNAs and represent almost 0.2% of cellular RNA pool. Early work focused on their roles in the nucleus, but more recently it has become clear that they also function in the cytoplasm, as reviewed here by Aillaud et al. In the cytoplasm, lncRNAs interact with ribosomes and regulate mRNA translation positively and negatively. They also interact with mitochondria and have roles in cell death and signal transduction during environmental changes, and also regulate endoplasmic reticulum (ER)- dependent autophagy. Various studies have shown the recruitment of lncRNAs with mRNAs in stress granules and with proteins in forming processing bodies. The authors also highlight the involvement of lncRNAs in ubiquitination-dependent protein processing and decay. This review is a great resource on how mammalian lncRNAs affect spatiotemporal responses of cytoplasmic organelles during stress condition. (Summary by Sunita Patak @psunita980) non-coding RNA 10.3390/ncrna6040044
ConnecTF: A platform to integrate transcription factor-gene interactions and validate regulatory networks
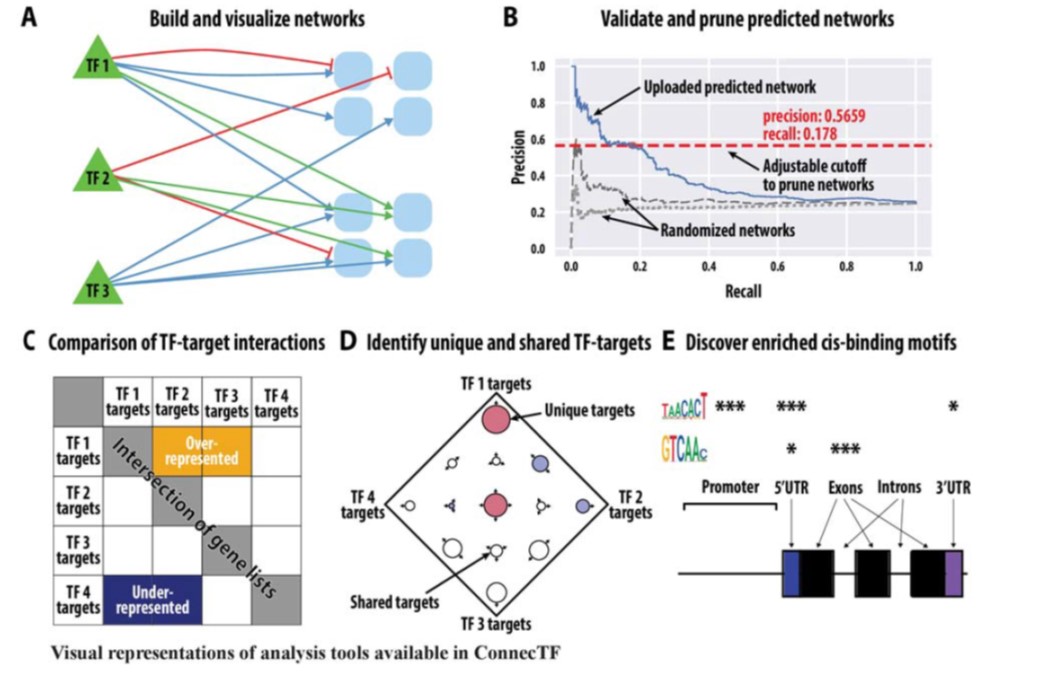 Gene regulatory networks (GFNs) are complex beasts, integrating multiple components of transcription factors (TFs), regulatory loci, and function — not to mention the myriad datasets and methodologies that generate datasets attempting to elucidate such networks. With rapid increases in technology to enable the discovery of more components within these networks, it is necessary to, in parallel, develop methods of organizing such large amounts of data in a user-friendly fashion. Such organizational tools are vital for compiling experimental data together towards the construction of robust GRNs. This was the mission of the authors in this study, who developed and released a web-based, species-independent platform called ConnecTF. ConnecTF, in their words, “integrates genome-wide studies of TF-target binding, TF-target regulation, and other TF-centric omic datasets and uses these to build and refine validated or inferred GRNs.” The authors demonstrate ConnecTF’s power in three case studies: first, identifying potential TF partners for certain TFs involved in ABA signaling; second, refining an inferred GRN for nitrogen signaling; and third, charting a full network path around NLP7, a master TF in nitrogen signaling. Overall, ConnecTF is ripe for use as a user-friendly, publicly available tool to compile and organize data for GRN construction and future GRN-related experiments. Check them out at https://ConnecTF.org! (Summary by Benjamin Jin) Plant Physiology 10.1093/plphys/kiaa012
Gene regulatory networks (GFNs) are complex beasts, integrating multiple components of transcription factors (TFs), regulatory loci, and function — not to mention the myriad datasets and methodologies that generate datasets attempting to elucidate such networks. With rapid increases in technology to enable the discovery of more components within these networks, it is necessary to, in parallel, develop methods of organizing such large amounts of data in a user-friendly fashion. Such organizational tools are vital for compiling experimental data together towards the construction of robust GRNs. This was the mission of the authors in this study, who developed and released a web-based, species-independent platform called ConnecTF. ConnecTF, in their words, “integrates genome-wide studies of TF-target binding, TF-target regulation, and other TF-centric omic datasets and uses these to build and refine validated or inferred GRNs.” The authors demonstrate ConnecTF’s power in three case studies: first, identifying potential TF partners for certain TFs involved in ABA signaling; second, refining an inferred GRN for nitrogen signaling; and third, charting a full network path around NLP7, a master TF in nitrogen signaling. Overall, ConnecTF is ripe for use as a user-friendly, publicly available tool to compile and organize data for GRN construction and future GRN-related experiments. Check them out at https://ConnecTF.org! (Summary by Benjamin Jin) Plant Physiology 10.1093/plphys/kiaa012
A network of transcriptional repressors modulates auxin responses
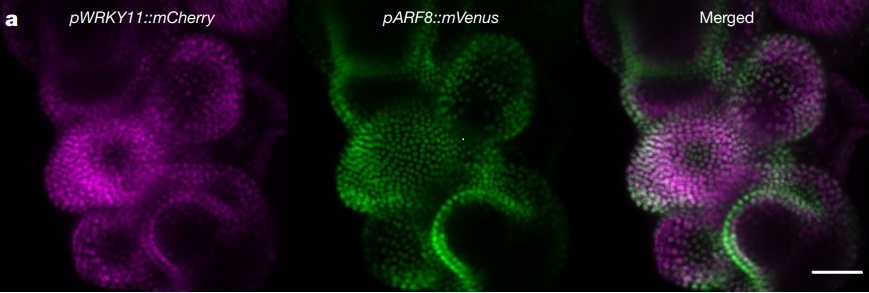
Untangling developmental programs and their methods of regulation are key to understanding the lives of all organisms. In plants, auxin is a hormone essential for plant growth and development. The class A AUSIN RESPONSE FACTOR (ARF) transcription factors necessary for auxin signaling throughout the plant lifecycle. While the expression patterns of these class A ARFs have been determined, how the individual signaling components are regulated as a system for development remains to be fully determined. In this study by Truskina et al., the authors perform multiple experiments to develop a model that implicates transcriptional repression as the main modulator of auxin responses, rather than a combination of chromatin modifications and transcriptional regulation. Their data first suggest that portions of chromatin coding for class A ARFs are constitutively open. Further, through an enhanced yeast one-hybrid screen, the authors identify a number of new transcription factors that bind to class A ARF genes, the majority of which demonstrate strong repressive activity. Expression and genetic analyses confirm these repressors’ involvement in the auxin regulatory network, and thus “reveal a functional regulatory network controlling the transcription of class A ARF genes… regulated by TF-mediated repression.” (Summary by Benjamin Jin) Nature 10.1038/s41586-020-2940-2
The canonical RdDM pathway mediates the control of seed germination timing under salinity ($)
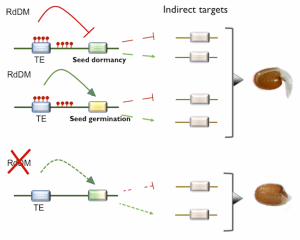 Epigenetic regulation can ensure that plant developmental programs and stress responses are tightly coordinated so that environmental changes do not compromise plant fitness. One of the mechanisms to achieve this is through RNA-directed DNA Methylation (RdDM), which is known to participate in several abiotic and biotic stress responses. In this research, Palomar and colleagues test the relative importance of RdDM in the germination of A. thaliana under high salinity conditions. Mutants for the core genes of the RdDM pathway had slower germination in saline conditions than the wild type, suggesting this mechanism is essential for plant responses to this stress. A detailed analysis of the dynamic of one of the core proteins –ARGONAUTE 4 (AGO4)– showed that its accumulation was temporally and spatially altered under high salinity. For instance, Whole-Genome Bisulfite Sequencing revealed that AGO4 modulates the methylation of genes that regulate seed germination and dormancy, such as EIN3 BINDING F-BOX1 and NAC4. Given this, the authors discuss that RdDM modulates seed germination in a stress context. As a result, this study expands our current knowledge about the role of the RdDM pathway in plant responses to abiotic stress in the earlier stages of their development. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant J. 10.1111/tpj.15064
Epigenetic regulation can ensure that plant developmental programs and stress responses are tightly coordinated so that environmental changes do not compromise plant fitness. One of the mechanisms to achieve this is through RNA-directed DNA Methylation (RdDM), which is known to participate in several abiotic and biotic stress responses. In this research, Palomar and colleagues test the relative importance of RdDM in the germination of A. thaliana under high salinity conditions. Mutants for the core genes of the RdDM pathway had slower germination in saline conditions than the wild type, suggesting this mechanism is essential for plant responses to this stress. A detailed analysis of the dynamic of one of the core proteins –ARGONAUTE 4 (AGO4)– showed that its accumulation was temporally and spatially altered under high salinity. For instance, Whole-Genome Bisulfite Sequencing revealed that AGO4 modulates the methylation of genes that regulate seed germination and dormancy, such as EIN3 BINDING F-BOX1 and NAC4. Given this, the authors discuss that RdDM modulates seed germination in a stress context. As a result, this study expands our current knowledge about the role of the RdDM pathway in plant responses to abiotic stress in the earlier stages of their development. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Plant J. 10.1111/tpj.15064
CLE40 induces localized root Ca2+ transients through CNGC9
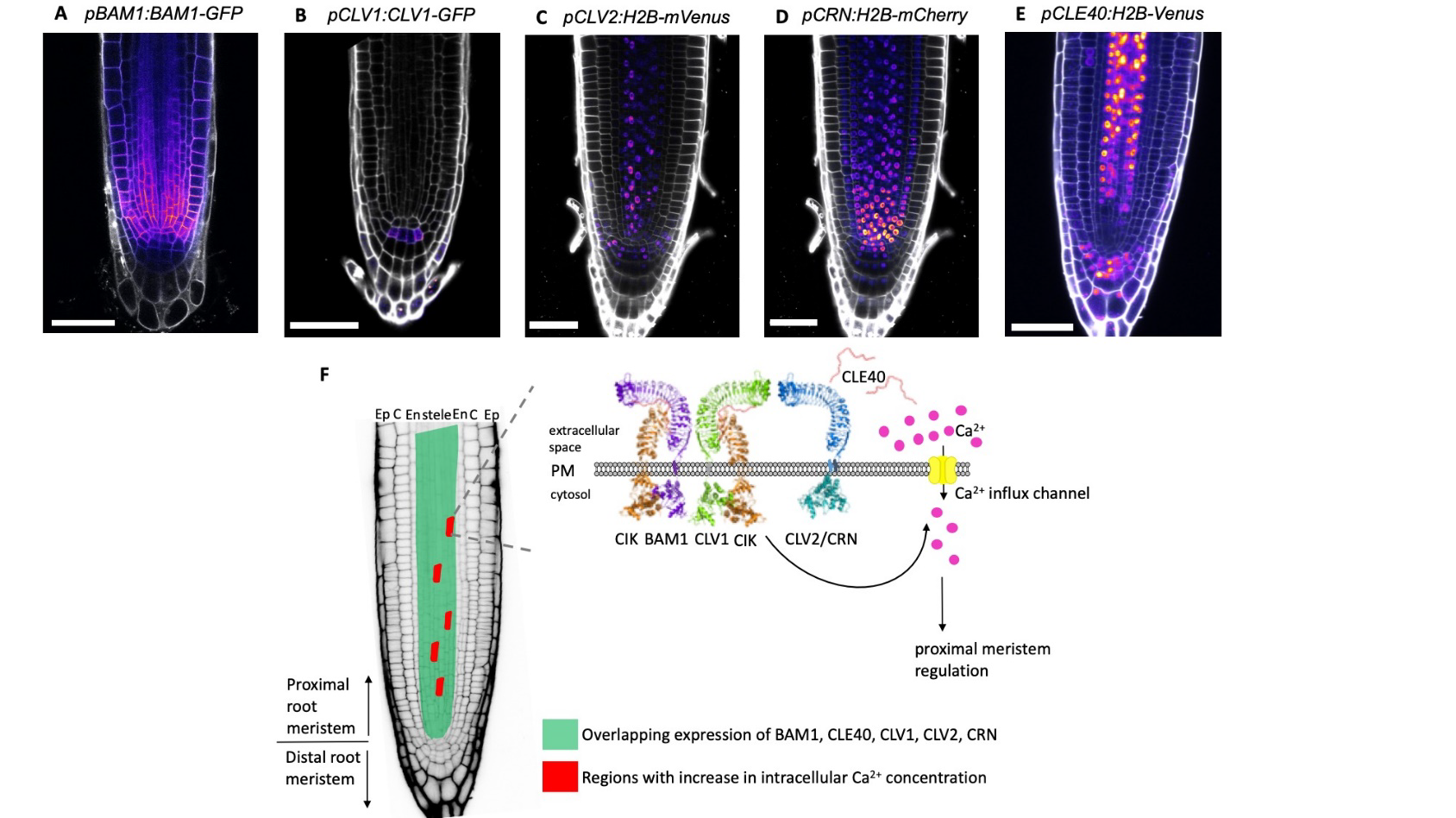 Growth and differentiation of cells in the meristematic region are tightly regulated by different signaling pathways. In the root meristem, the small peptide CLE40 acts as a ligand that binds to cell surface receptors and regulates cellular differentiation. In a new study, Breiden and colleagues have shown that application of external CLE40 peptide (CLE40p) induces highly localized Ca2+ transients in the proximal meristematic zone of the root. Application of a CLE40p with “randomized” amino acid sequence or application of CLE14p, a related peptide, does not elicit the same Ca2+ signals, suggesting a specific functional correlation. In agreement with their observation that the Ca2+ signals are sourced from the apoplast, the plasma membrane channel CNGC9 was seen to be required for CLE40p-induced transients. This Ca2+ signaling was also attenuated to varying degrees in mutants of receptors working downstream of CLE40 peptide signaling, suggesting their involvement in the signal transduction process. Though the authors have reasoned that it might be involved in regulation of cell fate, the precise role of this CLE40-induced Ca2+ signaling remains to be uncovered. (Summary by Pavithran Narayanan @pavi_narayanan) bioRxiv 10.1101/2020.11.13.381111v2
Growth and differentiation of cells in the meristematic region are tightly regulated by different signaling pathways. In the root meristem, the small peptide CLE40 acts as a ligand that binds to cell surface receptors and regulates cellular differentiation. In a new study, Breiden and colleagues have shown that application of external CLE40 peptide (CLE40p) induces highly localized Ca2+ transients in the proximal meristematic zone of the root. Application of a CLE40p with “randomized” amino acid sequence or application of CLE14p, a related peptide, does not elicit the same Ca2+ signals, suggesting a specific functional correlation. In agreement with their observation that the Ca2+ signals are sourced from the apoplast, the plasma membrane channel CNGC9 was seen to be required for CLE40p-induced transients. This Ca2+ signaling was also attenuated to varying degrees in mutants of receptors working downstream of CLE40 peptide signaling, suggesting their involvement in the signal transduction process. Though the authors have reasoned that it might be involved in regulation of cell fate, the precise role of this CLE40-induced Ca2+ signaling remains to be uncovered. (Summary by Pavithran Narayanan @pavi_narayanan) bioRxiv 10.1101/2020.11.13.381111v2
 Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis
Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis
Living organisms use biological clocks that are coordinated by temporal signals (time) and positional cues (space) during growth and development. In plants, the ‘root clock’ is generated by a mechanism involving rhythmic gene expression in a region called the oscillation zone (the border of the root elongation and differentiation zones) to produce lateral root primordia (LRP) approximately every 6 hours. Wachsman et al. identified the subcellular vesicle trafficking and pectin components as part of the root clock regulators and LRP initiation factors. The LRP cell initiation starts soon after formation of a prebranch site. By dividing the root into several positions, collecting root sections from different times, and analysing them by RNA-sequencing, the authors identified cell wall, vesicle trafficking, and membrane-related genes required for the LRP formation. Mutagenized DR5::LUC seeds screening found the vesicle-trafficking gene GNOM and its suppressor, AGD3, as a novel root clock regulators. GNOM is required for pectin distribution at the cell wall which is synthesized in its esterified form in the Golgi. At the sites of LRP initiation, the pectin subtypes are differentially distributed. This finding uncovers novel components that are essential for root clock regulation and lateral root initiation (Summary by Min May Wong @wongminmay) Science 10.1126/science.abb7250
Microbiota-root-shoot axis modulation by MYC2 favors Arabidopsis growth over defense under suboptimal light
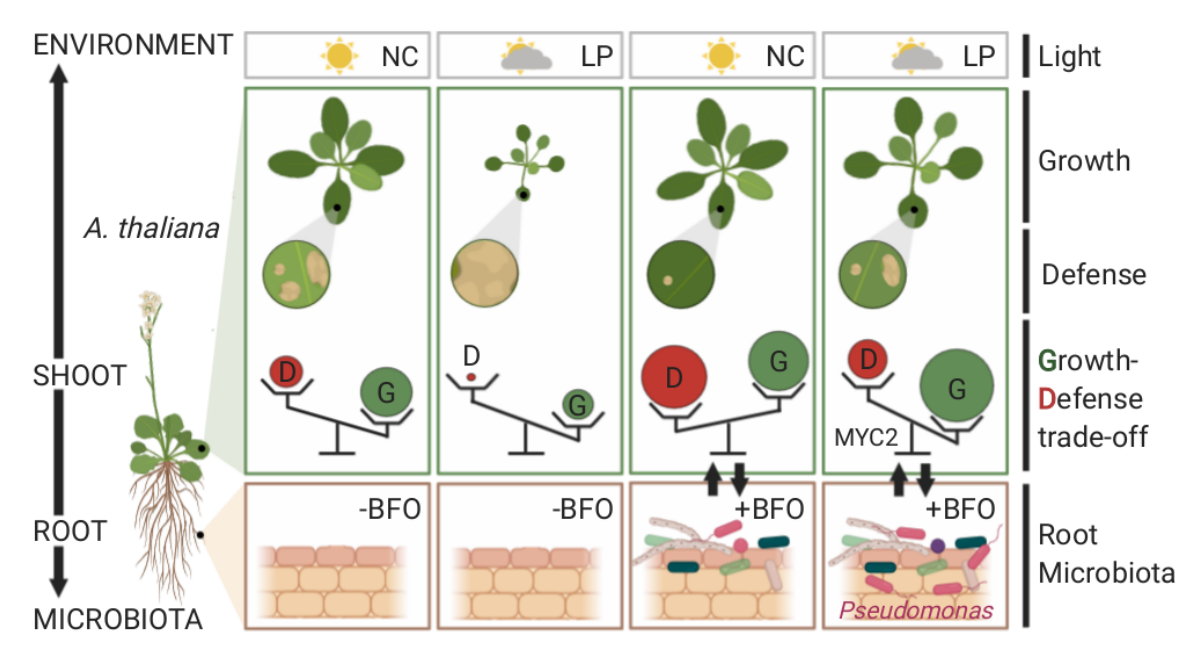 Below- and aboveground plant organs experience distinct biotic and abiotic environments. Thus, coordination between root and shoot responses are likely crucial for plant survival. Given that a substantial amount of photosynthetically fixed carbons is invested in the rhizosphere, Hou et al. hypothesized that the aboveground responses to light and the belowground responses to microbes are interconnected. Using a gnotobiotic system and synthetic microbial communities (bacteria, fungi, and oomycetes), the authors showed that aboveground light conditions alter Arabidopsis root microbiota assembly, and the microbiota can help plants grow under a reduced light condition. Transcriptome analyses revealed that the microbiota influences shoot responses to light, whereas the light condition modulates root responses to microbes. Such microbiota-root-shoot axis connection might allow plants to prioritize growth over defense under low light. While plant responses were steered towards growth over defense, the microbiota could protect leaves from pathogen attack under the suboptimal light condition. The authors identified MYC2, a master regulator of jasmonic acid signaling, as a key plant factor that governs microbiota-mediated plant growth promotion and protection from pathogens under low light. This study suggests the potential role of bidirectional root-shoot signaling in orchestrating a light-dependent growth-defense trade-off. (Summary by Tatsuya Nobori @nobolly) bioRxiv https://doi.org/10.1101/2020.11.06.371146
Below- and aboveground plant organs experience distinct biotic and abiotic environments. Thus, coordination between root and shoot responses are likely crucial for plant survival. Given that a substantial amount of photosynthetically fixed carbons is invested in the rhizosphere, Hou et al. hypothesized that the aboveground responses to light and the belowground responses to microbes are interconnected. Using a gnotobiotic system and synthetic microbial communities (bacteria, fungi, and oomycetes), the authors showed that aboveground light conditions alter Arabidopsis root microbiota assembly, and the microbiota can help plants grow under a reduced light condition. Transcriptome analyses revealed that the microbiota influences shoot responses to light, whereas the light condition modulates root responses to microbes. Such microbiota-root-shoot axis connection might allow plants to prioritize growth over defense under low light. While plant responses were steered towards growth over defense, the microbiota could protect leaves from pathogen attack under the suboptimal light condition. The authors identified MYC2, a master regulator of jasmonic acid signaling, as a key plant factor that governs microbiota-mediated plant growth promotion and protection from pathogens under low light. This study suggests the potential role of bidirectional root-shoot signaling in orchestrating a light-dependent growth-defense trade-off. (Summary by Tatsuya Nobori @nobolly) bioRxiv https://doi.org/10.1101/2020.11.06.371146



