Plant Science Research Weekly: December 15, 2023
Special issue: Human-machine collaboration in plant biology
![]() This is an excellent article to wrap up this year and lead us into the future. Introducing a special issue of Plant Cell Physiology, Nakajima et al. summarize an exciting collection of papers that look at diverse ways that plant biology can be enhanced through “human-machine collaborations”. Some of these, like microscopes (machines that enhance human vision) are already widely used, whereas others have a futuristic aura, such as P-MIRU (Polarization Multispectral Imaging for Reflection from Biological Surfaces). Many of the approaches use trainable algorithms such as artificial intelligence (AI), explainable AI, and deep-learning to provide insights into plant development and physiology that go way beyond human capabilities. Examples include methods that track and trace cells in 4D (at the root tip, cotyledons, and zygote). Another useful machine is a portable tool for quantifying stomatal movements in intact leaves, greatly increasing the rate at which data can be collected. Several articles describe how machines can be trained to interpret huge sets of data and images, leading to observations beyond human capabilities. This is an exciting article to share with your students. (Summary by Mary Williams @PlantTeaching) Plant Cell Physiol. 10.1093/pcp/pcad144
This is an excellent article to wrap up this year and lead us into the future. Introducing a special issue of Plant Cell Physiology, Nakajima et al. summarize an exciting collection of papers that look at diverse ways that plant biology can be enhanced through “human-machine collaborations”. Some of these, like microscopes (machines that enhance human vision) are already widely used, whereas others have a futuristic aura, such as P-MIRU (Polarization Multispectral Imaging for Reflection from Biological Surfaces). Many of the approaches use trainable algorithms such as artificial intelligence (AI), explainable AI, and deep-learning to provide insights into plant development and physiology that go way beyond human capabilities. Examples include methods that track and trace cells in 4D (at the root tip, cotyledons, and zygote). Another useful machine is a portable tool for quantifying stomatal movements in intact leaves, greatly increasing the rate at which data can be collected. Several articles describe how machines can be trained to interpret huge sets of data and images, leading to observations beyond human capabilities. This is an exciting article to share with your students. (Summary by Mary Williams @PlantTeaching) Plant Cell Physiol. 10.1093/pcp/pcad144
A novel way to conduct genome wide association studies for secondary metabolites
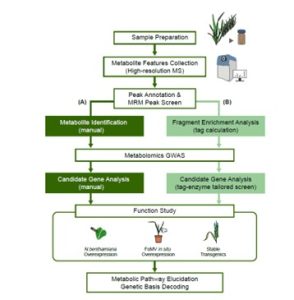 Plants produce many specialized metabolites that are often associated with increased plant fitness. Metabolic genome wide association studies (mGWAS) identify enzymes involved in specialized metabolite biosynthesis. However, this relies on identifying metabolites from mass spectrometry data which is often time and labor intensive. Here, Zhu et al. bypassed this step by identifying structural modifications rather than the entire metabolite. They used liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) on leaf extracts from 391 diverse wheat accessions and identified six modifications in the metabolites: hydration, glycosylation, methylation, acetylation, decarboxylation and acylation. This information was fed into a mGWAS pipeline that identified 3594 loci associated with the modifications. To reduce this number, they excluded loci that did not contain an enzyme capable of performing the modification and loci that did not have single nucleotide polymorphisms (SNPs) in the coding sequence or promoter of this gene. This successfully reduced the 3594 loci to 499 candidates. The authors showed that the function of these candidates can be validated in wheat using a Foxtail mosaic virus transient expression system. This strategy is a vast improvement on the traditional mGWAS method as it removes the time-consuming step of metabolite identification and is an unbiased way of identifying novel gene candidates. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad286
Plants produce many specialized metabolites that are often associated with increased plant fitness. Metabolic genome wide association studies (mGWAS) identify enzymes involved in specialized metabolite biosynthesis. However, this relies on identifying metabolites from mass spectrometry data which is often time and labor intensive. Here, Zhu et al. bypassed this step by identifying structural modifications rather than the entire metabolite. They used liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) on leaf extracts from 391 diverse wheat accessions and identified six modifications in the metabolites: hydration, glycosylation, methylation, acetylation, decarboxylation and acylation. This information was fed into a mGWAS pipeline that identified 3594 loci associated with the modifications. To reduce this number, they excluded loci that did not contain an enzyme capable of performing the modification and loci that did not have single nucleotide polymorphisms (SNPs) in the coding sequence or promoter of this gene. This successfully reduced the 3594 loci to 499 candidates. The authors showed that the function of these candidates can be validated in wheat using a Foxtail mosaic virus transient expression system. This strategy is a vast improvement on the traditional mGWAS method as it removes the time-consuming step of metabolite identification and is an unbiased way of identifying novel gene candidates. (Summary by Rose McNelly @Rose_McN) Plant Cell 10.1093/plcell/koad286
Breakthrough in the identification of photosynthesis genes in green algae
 Imagine the everyday delights of foods like bread, ramen, or sushi. Central to these culinary staples is starch, a product of the photosynthesis process in plants. Despite its critical role, people still do not fully understand the regulation and biogenesis of the photosynthesis machinery. In this study, Kafri et al. identified 70 previously unknown photosynthesis genes in the green algae species Chlamydomonas reinhardtii, offering deeper understanding of this process. Both wild-type and photosynthesis-deficient Chlamydomonas can grow in dark environments when supplied with acetate as a carbon source, but in light conditions without acetate, photosynthesis-deficient mutants display notable growth defects. Leveraging this characteristic, the authors conducted a high-throughput automated spot test on agar to verify a set of 1,781 previously identified photosynthesis-deficient Chlamydomonas mutants. Subsequently, the authors designed a pooled backcrossing strategy to map the causal mutations and narrowed the pool down to 115 high fidelity genes, including 70 previously uncharacterized genes. To validate their findings, the team performed genetic complementation for some of the hits and examined their protein localization. Further, a large-scale proteomic profiling of these mutants informed the impact of the loss of each gene on the proteome, with some causing strong depletion of the photosynthetic complexes, indicating their regulatory roles. Notably, two master regulators of photosynthesis, MTF1 and PMR1, were identified. This study marks a significant leap forward for the understanding of the photosynthetic machinery in Chlamydomonas, and provides an important resource to pave the way for further explorations of photosynthesis. (Summary by Xiaohui Li @Xiao_hui_Li) Cell 10.1016/j.cell.2023.11.007
Imagine the everyday delights of foods like bread, ramen, or sushi. Central to these culinary staples is starch, a product of the photosynthesis process in plants. Despite its critical role, people still do not fully understand the regulation and biogenesis of the photosynthesis machinery. In this study, Kafri et al. identified 70 previously unknown photosynthesis genes in the green algae species Chlamydomonas reinhardtii, offering deeper understanding of this process. Both wild-type and photosynthesis-deficient Chlamydomonas can grow in dark environments when supplied with acetate as a carbon source, but in light conditions without acetate, photosynthesis-deficient mutants display notable growth defects. Leveraging this characteristic, the authors conducted a high-throughput automated spot test on agar to verify a set of 1,781 previously identified photosynthesis-deficient Chlamydomonas mutants. Subsequently, the authors designed a pooled backcrossing strategy to map the causal mutations and narrowed the pool down to 115 high fidelity genes, including 70 previously uncharacterized genes. To validate their findings, the team performed genetic complementation for some of the hits and examined their protein localization. Further, a large-scale proteomic profiling of these mutants informed the impact of the loss of each gene on the proteome, with some causing strong depletion of the photosynthetic complexes, indicating their regulatory roles. Notably, two master regulators of photosynthesis, MTF1 and PMR1, were identified. This study marks a significant leap forward for the understanding of the photosynthetic machinery in Chlamydomonas, and provides an important resource to pave the way for further explorations of photosynthesis. (Summary by Xiaohui Li @Xiao_hui_Li) Cell 10.1016/j.cell.2023.11.007
Guanylate cyclase activity of TIR1/AFB auxin receptors in rapid auxin responses
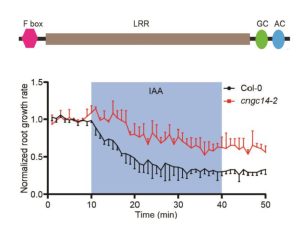 Auxin is a pleiotropic plant hormone with diverse functions, many of which are mediated through transcriptional reprograming. However, some auxin responses occur extremely rapidly, ruling out changes in transcription as a mechanism. In 2022, one of the components of an auxin receptor, F-box protein TIR1/AFB, was discovered to include an adenylate cyclase catalytic domain, suggesting a role for cAMP signaling. However, this finding didn’t explain all of the observed effects, particularly the rapid calcium oscillations induced by auxin. Here, Qi et al. provide evidence for a guanylate cyclase catalytic domain adjacent to the previously identified adenylate cyclase domain. They found that cGMP production is rapidly stimulated by auxin, and is involved in the rapid calcium oscillations and root growth responses. These results demonstrate that cGMP is an important second messenger in the auxin response. (Summary by Mary Williams @PlantTeaching) bioRxiv https://www.biorxiv.org/content/10.1101/2023.11.18.567481v1
Auxin is a pleiotropic plant hormone with diverse functions, many of which are mediated through transcriptional reprograming. However, some auxin responses occur extremely rapidly, ruling out changes in transcription as a mechanism. In 2022, one of the components of an auxin receptor, F-box protein TIR1/AFB, was discovered to include an adenylate cyclase catalytic domain, suggesting a role for cAMP signaling. However, this finding didn’t explain all of the observed effects, particularly the rapid calcium oscillations induced by auxin. Here, Qi et al. provide evidence for a guanylate cyclase catalytic domain adjacent to the previously identified adenylate cyclase domain. They found that cGMP production is rapidly stimulated by auxin, and is involved in the rapid calcium oscillations and root growth responses. These results demonstrate that cGMP is an important second messenger in the auxin response. (Summary by Mary Williams @PlantTeaching) bioRxiv https://www.biorxiv.org/content/10.1101/2023.11.18.567481v1
Tree or bush? It’s all in the hormones
 Much of our understanding of the molecular underpinning that control shoot architecture comes from studies of annual plants such as Arabidopsis, pea, and rice. This new work by Su et al. investigates shoot branching in a long-lived tree, silver birch (Betula pendula). They started with a naturally occurring bushy variant, which has a premature stop codon in a gene encoding a strigolactone (SL) biosynthetic enzyme, BpMAX1, and correspondingly low levels of SL. The bushiness of the SL-deficient mutants is due to a shorter stem along with increased outgrowth of higher-order branches; 13-year old mutant trees form up to seventh-order branches, while the wild-type trees max out at fourth-order branches. The authors then looked at auxin transport and auxin levels in the mutant and found that polar-auxin transport is unaffected. However, in wild-type plants there is an auxin gradient from tip to base, which is not observed in the bushy mutants. They used mathematical modeling to investigate the origin of this difference and concluded that the bushiness itself (shorter internodes and more growing tips) is sufficient to explain the increased auxin levels at the base, “with no requirement for differences in auxin synthesis, transport, or decay.” As the authors observe, understanding how branching is controlled in trees has a potential for economic impact, with tall unbranched trees being preferred for timber, and shorter branchier trees being preferred for fruit production. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2308587120
Much of our understanding of the molecular underpinning that control shoot architecture comes from studies of annual plants such as Arabidopsis, pea, and rice. This new work by Su et al. investigates shoot branching in a long-lived tree, silver birch (Betula pendula). They started with a naturally occurring bushy variant, which has a premature stop codon in a gene encoding a strigolactone (SL) biosynthetic enzyme, BpMAX1, and correspondingly low levels of SL. The bushiness of the SL-deficient mutants is due to a shorter stem along with increased outgrowth of higher-order branches; 13-year old mutant trees form up to seventh-order branches, while the wild-type trees max out at fourth-order branches. The authors then looked at auxin transport and auxin levels in the mutant and found that polar-auxin transport is unaffected. However, in wild-type plants there is an auxin gradient from tip to base, which is not observed in the bushy mutants. They used mathematical modeling to investigate the origin of this difference and concluded that the bushiness itself (shorter internodes and more growing tips) is sufficient to explain the increased auxin levels at the base, “with no requirement for differences in auxin synthesis, transport, or decay.” As the authors observe, understanding how branching is controlled in trees has a potential for economic impact, with tall unbranched trees being preferred for timber, and shorter branchier trees being preferred for fruit production. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2308587120
Diurnal switches in diazotrophic lifestyle increase nitrogen contribution to cereals
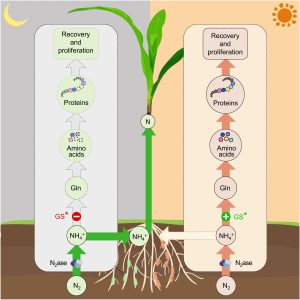 Unlike legumes, which form symbiotic associations with nitrogen-fixing bacteria (diazotrophs), high-yielding cereal crops are usually supplemented with inorganic fertilizers that are both energetically and environmentally problematic. Some non-legumes can obtain nitrogen from associations with free-living diazotrophs, and several strategies are being pursued to optimize such associations. A major challenge is that in most of these conditions the fixed nitrogen is assimilated into glutamine (Gln) by the glutamine synthase (GS) enzyme in the prokaryote, switching off the nitrogen assimilation pathway and retaining most of the fixed nitrogen in the bacterial cell, providing little to the plant. Efforts to modify this pathway leads to poor growth in the bacteria and a high rate of phenotypic reversion. Here, Tang et al. have exploited a mutation in the GS gene that makes the encoded enzyme less sensitive to negative feedback but also cold sensitive, with low activity levels at 23 °C. When this mutation was introduced into the diazotroph Klebsiella oxytoca grown in association with rice or maize, it led to a diurnal pattern of GS activity driven by temperature cycling. During the cooler nighttime, nitrogen fixed by the bacteria is excreted and available to the plant, but during the warmer daytime the active GS enzyme retains the nitrogen in the prokaryotic cell. This diurnal cycling allows the prokaryote time to recover and proliferate, improving nitrogen donation to the crop. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-023-43370-4
Unlike legumes, which form symbiotic associations with nitrogen-fixing bacteria (diazotrophs), high-yielding cereal crops are usually supplemented with inorganic fertilizers that are both energetically and environmentally problematic. Some non-legumes can obtain nitrogen from associations with free-living diazotrophs, and several strategies are being pursued to optimize such associations. A major challenge is that in most of these conditions the fixed nitrogen is assimilated into glutamine (Gln) by the glutamine synthase (GS) enzyme in the prokaryote, switching off the nitrogen assimilation pathway and retaining most of the fixed nitrogen in the bacterial cell, providing little to the plant. Efforts to modify this pathway leads to poor growth in the bacteria and a high rate of phenotypic reversion. Here, Tang et al. have exploited a mutation in the GS gene that makes the encoded enzyme less sensitive to negative feedback but also cold sensitive, with low activity levels at 23 °C. When this mutation was introduced into the diazotroph Klebsiella oxytoca grown in association with rice or maize, it led to a diurnal pattern of GS activity driven by temperature cycling. During the cooler nighttime, nitrogen fixed by the bacteria is excreted and available to the plant, but during the warmer daytime the active GS enzyme retains the nitrogen in the prokaryotic cell. This diurnal cycling allows the prokaryote time to recover and proliferate, improving nitrogen donation to the crop. (Summary by Mary Williams @PlantTeaching) Nature Comms. 10.1038/s41467-023-43370-4
Engineering wheat to reduce the immunoreactivity of gluten
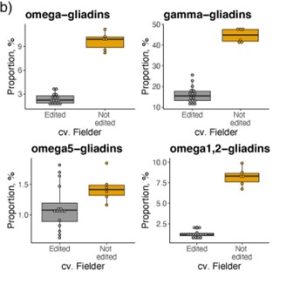 Some people cannot eat wheat because they are sensitive to gluten, which is a proteinaceous network of glutenins and gliadins. Gliadins are divided into four classes, ω, γ, α, and β, with ω- and γ-gliadins being the most toxic to gluten sensitive individuals. Hence, there is interest in reducing ω- and γ-gliadin content. Here Yu et al. annotated 9 functional ω-gliadin and 22 functional γ-gliadin genes in the genome of the wheat cultivar Fielder. Using these annotations, they designed seven guide RNAs (gRNAs) to target all the ω-gliadin genes and some of the γ-gliadin genes. The gRNAs and a Cas9 construct were transformed into Fielder by biolistic transformation. Whole genome sequencing identified a transgene free wheat line with deletions in all ω-gliadin genes and 55% of the γ-gliadin genes. This caused a 75% reduction in ω-gliadin and a 64% reduction in γ-gliadin protein abundance. More importantly, this led to a 42-47 fold reduction in immunoreactivity to antibodies which recognise toxic epitopes. This does not quite reach levels to be consumed by those who are gluten sensitive, but reducing ω- and γ-gliadin content even further could be a promising strategy for generating wheat that is safe for gluten sensitive individuals. (Summary by Rose McNelly @Rose_McN) Plant Biotech. J. 10.1111/pbi.14231
Some people cannot eat wheat because they are sensitive to gluten, which is a proteinaceous network of glutenins and gliadins. Gliadins are divided into four classes, ω, γ, α, and β, with ω- and γ-gliadins being the most toxic to gluten sensitive individuals. Hence, there is interest in reducing ω- and γ-gliadin content. Here Yu et al. annotated 9 functional ω-gliadin and 22 functional γ-gliadin genes in the genome of the wheat cultivar Fielder. Using these annotations, they designed seven guide RNAs (gRNAs) to target all the ω-gliadin genes and some of the γ-gliadin genes. The gRNAs and a Cas9 construct were transformed into Fielder by biolistic transformation. Whole genome sequencing identified a transgene free wheat line with deletions in all ω-gliadin genes and 55% of the γ-gliadin genes. This caused a 75% reduction in ω-gliadin and a 64% reduction in γ-gliadin protein abundance. More importantly, this led to a 42-47 fold reduction in immunoreactivity to antibodies which recognise toxic epitopes. This does not quite reach levels to be consumed by those who are gluten sensitive, but reducing ω- and γ-gliadin content even further could be a promising strategy for generating wheat that is safe for gluten sensitive individuals. (Summary by Rose McNelly @Rose_McN) Plant Biotech. J. 10.1111/pbi.14231
Volatile methyl jasmonate from roots triggers host-beneficial soil microbiome biofilms
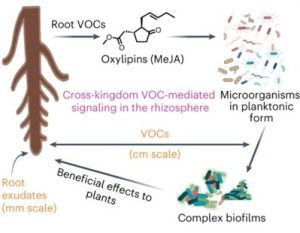 Volatile molecules released from plant roots (root VOCs, rVOCs) can diffuse over long distances, thereby increasing the sphere of plant influence. However, their influence on complex soil microbial communities (or microbiomes) and ecological implications are poorly understood, mainly due to technical challenges in capturing and quantifying rVOCs. To overcome these challenges, Kulkarni and Mazumder et al. developed an airflow system named the “push-pull system” to investigate how rVOCs affect biofilm formation in soil microbiomes. They established that rVOCs trigger lifestyle transition from planktonic to biofilm states in complex soil microbial communities. The authors showed that this phenomenon is evolutionarily conserved in plant kingdom, ranging from ferns to higher plants. By volatile profiling of biosynthetic mutant lines, they identified methyl jasmonate (MeJA) as the principal bioactive compound in rVOCs and established that MeJA acts at nanomolar concentrations as a signal responsible for increasing biofilm formation by soil microbiomes. The authors also showed that the MeJA-induced biofilms promote plant growth from a distance, through microbial volatile signaling. Thus, a volatile-mediated feedback loop was established between plant and complex soil microbial communities. This study (i) adds evidence to the emerging concept of ‘extended rhizosphere’, through volatile-mediated long-distance influence, and (ii) discovers a moonlighting role of jasmonate, where the host defense signal MeJA is co-opted by plant roots for assembling host-beneficial biofilms in the soil microbiome. (Summary by Arijit Mukherjee @ArijitM61745830) Nat. Chem. Biol. 10.1038/s41589-023-01462-8
Volatile molecules released from plant roots (root VOCs, rVOCs) can diffuse over long distances, thereby increasing the sphere of plant influence. However, their influence on complex soil microbial communities (or microbiomes) and ecological implications are poorly understood, mainly due to technical challenges in capturing and quantifying rVOCs. To overcome these challenges, Kulkarni and Mazumder et al. developed an airflow system named the “push-pull system” to investigate how rVOCs affect biofilm formation in soil microbiomes. They established that rVOCs trigger lifestyle transition from planktonic to biofilm states in complex soil microbial communities. The authors showed that this phenomenon is evolutionarily conserved in plant kingdom, ranging from ferns to higher plants. By volatile profiling of biosynthetic mutant lines, they identified methyl jasmonate (MeJA) as the principal bioactive compound in rVOCs and established that MeJA acts at nanomolar concentrations as a signal responsible for increasing biofilm formation by soil microbiomes. The authors also showed that the MeJA-induced biofilms promote plant growth from a distance, through microbial volatile signaling. Thus, a volatile-mediated feedback loop was established between plant and complex soil microbial communities. This study (i) adds evidence to the emerging concept of ‘extended rhizosphere’, through volatile-mediated long-distance influence, and (ii) discovers a moonlighting role of jasmonate, where the host defense signal MeJA is co-opted by plant roots for assembling host-beneficial biofilms in the soil microbiome. (Summary by Arijit Mukherjee @ArijitM61745830) Nat. Chem. Biol. 10.1038/s41589-023-01462-8
Old reserves and ancient buds fuel regrowth of coast redwood after catastrophic fire
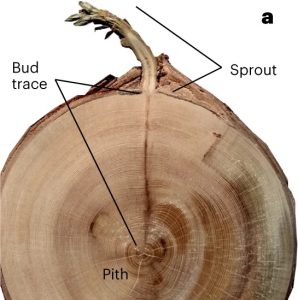 In recent years we have witnessed catastrophic fires throughout the world, including the redwood forests of California. Although these huge ancient trees are fire resistant, many have died due to the extreme heat generated by these recent fires. Here, Peltier et al. examined patterns of regrowth from burned trees. In a series of elegant studies, they revealed the remarkable resilience of these treasures of the plant kingdom. First, using carbon dating, they examined the age of the carbohydrate incorporated into the regenerating tissues, and found that some had been stored in the trees for decades. Second, they looked at the bud sources for regenerating shoots. By tracing the journey of the bud outgrowth through the tree rings, they were able to demonstrate that some of the buds were formed hundreds of years ago and lay dormant until activated. These results show the remarkable ability of these long-lived trees to survive, raising hope that they’ll persist after the end of the Anthropocene. However, it’s not a sure thing. As the authors observe, as these superannuated life-giving reservoirs are depleted, what will happen after the next catastrophic fire? (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-023-01581-z
In recent years we have witnessed catastrophic fires throughout the world, including the redwood forests of California. Although these huge ancient trees are fire resistant, many have died due to the extreme heat generated by these recent fires. Here, Peltier et al. examined patterns of regrowth from burned trees. In a series of elegant studies, they revealed the remarkable resilience of these treasures of the plant kingdom. First, using carbon dating, they examined the age of the carbohydrate incorporated into the regenerating tissues, and found that some had been stored in the trees for decades. Second, they looked at the bud sources for regenerating shoots. By tracing the journey of the bud outgrowth through the tree rings, they were able to demonstrate that some of the buds were formed hundreds of years ago and lay dormant until activated. These results show the remarkable ability of these long-lived trees to survive, raising hope that they’ll persist after the end of the Anthropocene. However, it’s not a sure thing. As the authors observe, as these superannuated life-giving reservoirs are depleted, what will happen after the next catastrophic fire? (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-023-01581-z
That’s it for 2023, we’ll be back in 2024. Until then, Happy New Year from the Plant Science Research Weekly team, thanks for reading!